The goal of magrene is to identify and analyze graph motifs containing duplicated genes in gene regulatory networks (GRNs). Possible motifs include V, PPI V, lambda, delta, and bifans.
Motif frequencies can be analyzed in the context of gene duplications to explore the impact of small-scale and whole-genome duplications on gene regulatory networks.
Additionally, GRNs can be tested for motif enrichment by comparing motif frequencies to a null distribution generated from degree-preserving simulated GRNs.
Finally, users can calculate the interaction similarity between gene pairs based on the Sorensen-Dice similarity index.
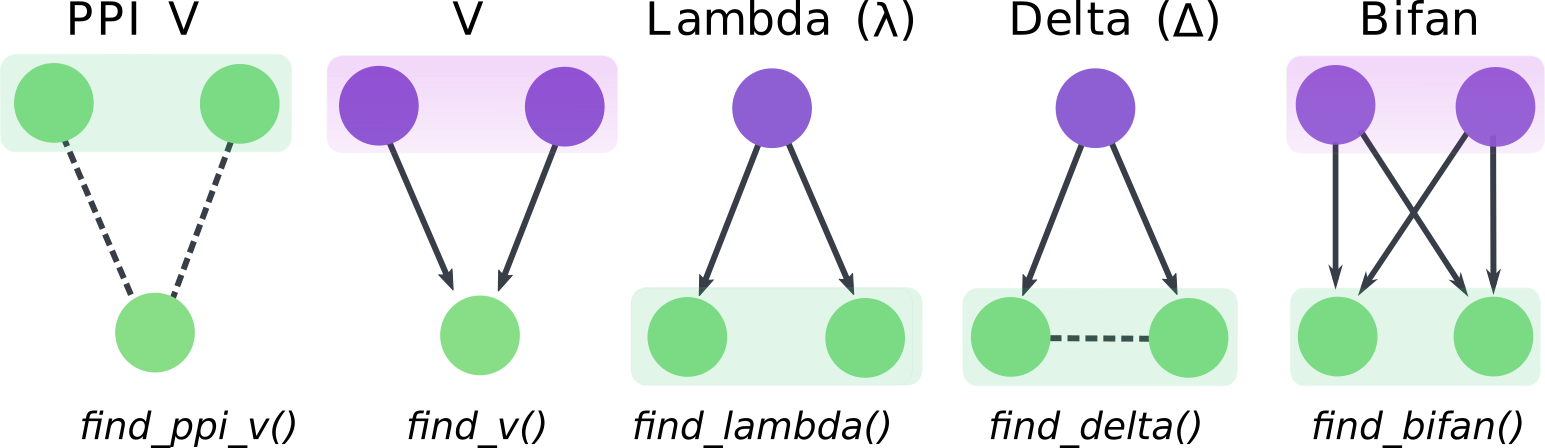
Network motifs and functions to identify them. Shaded boxes indicate paralogs. Regulators and targets are indicated in purple and green, respectively. Arrows indicate directed regulatory interactions, while dashed lines indicate protein-protein interaction.
Installation instructions
Get the latest stable R release from CRAN. Then install magrene using from Bioconductor the following code:
if (!requireNamespace("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
BiocManager::install("magrene")And the development version from GitHub with:
BiocManager::install("almeidasilvaf/magrene")Citation
Below is the citation output from using citation('magrene') in R. Please run this yourself to check for any updates on how to cite magrene.
print(citation('magrene'), bibtex = TRUE)
#>
#> To cite package 'magrene' in publications use:
#>
#> Almeida-Silva F, Van de Peer Y (2022). _magrene: Motif Analysis In
#> Gene Regulatory Networks_. R package version 0.99.0,
#> <https://github.com/almeidasilvaf/magrene>.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {magrene: Motif Analysis In Gene Regulatory Networks},
#> author = {Fabrício Almeida-Silva and Yves {Van de Peer}},
#> year = {2022},
#> note = {R package version 0.99.0},
#> url = {https://github.com/almeidasilvaf/magrene},
#> }Please note that magrene was only made possible thanks to many other R and bioinformatics software authors, which are cited either in the vignettes and/or the paper(s) describing this package.
Code of Conduct
Please note that the magrene project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
Development tools
- Continuous code testing is possible thanks to GitHub actions through usethis, remotes, and rcmdcheck customized to use Bioconductor’s docker containers and BiocCheck.
- Code coverage assessment is possible thanks to codecov and covr.
- The documentation website is automatically updated thanks to pkgdown.
- The documentation is formatted thanks to devtools and roxygen2.
For more details, check the dev directory.
This package was developed using biocthis.
