set.seed(123) # for reproducibility
# Load required packages
library(STRINGdb)
library(igraph)
library(tidyverse)4 Analysis of protein-protein interaction (PPI) networks
In this lesson, you will learn how to obtain protein-protein interaction (PPI) data and analyze PPI networks. At the end of this lesson, you will be able to:
- obtain PPI networks from the STRING database (Szklarczyk et al. 2023);
- identify clusters in PPI networks;
- perform a functional profiling of subgraphs;
- explore and visualize subgraphs
Let’s start by loading the packages we will use.
4.1 Getting to know the example data
Here, we will use an example data set named diff_exp_example1, available in STRINGdb. Let’s first load it and take a quick look at it.
# Load the example data set
data(diff_exp_example1)
head(diff_exp_example1) pvalue logFC gene
1 0.0001018 3.333461 VSTM2L
2 0.0001392 3.822383 TBC1D2
3 0.0001720 3.306056 LENG9
4 0.0001739 3.024605 TMEM27
5 0.0001990 3.854414 LOC100506014
6 0.0002393 3.082052 TSPAN1This data frame contains gene-wise test statistics for a differential expression analysis (i.e., log fold changes and P-values) obtained with BiocStyle::Biocpkg("limma"). In the original study, authors compared gene expression levels of A549 lung cancer cells 48 hours after exposure to Resveratrol (a natural phytoestrogen found in red wine and other plants) with control conditions (ethanol only). Genes are sorted by FDR-corrected P-values.
4.2 Obtaining data from STRING
The STRING database is the largest database of known and predicted protein-protein interactions, and it contains both direct (physical) and indirect (functional) associations. The STRINGdb package provides users with a friendly interface to the STRING database. This package uses an object-oriented approach, so all computations can be performed by calling methods associated with an object of class STRINGdb. The STRINGdb object contains a connection to the STRING database, and it can be created as follows:
string_db <- STRINGdb$new(
1 version = "12.0",
2 species = 9606,
3 score_threshold = 400,
4 network_type = "full"
)- 1
- STRING version to be used.
- 2
- NCBI Taxonomy ID of the species to be used (here, Homo sapiens)
- 3
- Minimum score (scale from 0 to 1000)
- 4
- Network type (here, ‘full’). Other options include ‘functional’ (only functional interactions), and ‘physical’ (only physical interactions).
To see all possible arguments of the new() method, you can use:
STRINGdb$help()Usage: $help(topic) where topic is the name of a method (quoted or not)
The definition of class STRINGdb follows.
Reference Class "STRINGdb":
Class fields:
Name: annotations annotations_description
Class: data.frame data.frame
Name: graph homology_graph
Class: igraph igraph
Name: proteins aliases_tf
Class: data.frame data.frame
Name: aliases_type species
Class: character numeric
Name: version network_type
Class: character character
Name: input_directory backgroundV
Class: character vector
Name: score_threshold pathways_benchmark_blackList
Class: numeric data.frame
Name: stable_url protocol
Class: character character
Name: file_version
Class: character
Class Methods:
"get_link", "get_annotations", "get_pathways_benchmarking_blackList",
"ppi_enrichment", "export", "add_diff_exp_color",
"add_proteins_description", "callSuper", "get_aliases",
"get_term_proteins", ".objectParent", "benchmark_ppi", "get_proteins",
"get_ppi_enrichment", "get_ppi_enrichment_full", "get_graph", "untrace",
"get_pubmed_interaction", "get_homologs", "get_paralogs", "load",
"enrichment_heatmap", "trace", ".objectPackage", "get_neighbors",
"getClass", "get_homology_graph", "get_bioc_graph", "import",
"get_summary", "copy", "getRefClass", "get_interactions",
"set_background", "get_subnetwork", "field", "get_homologs_besthits",
"map", "benchmark_ppi_pathway_view", "plot_ppi_enrichment",
"get_enrichment", "load_all", "mp", "usingMethods", "plot_network",
"initialize", "initFields", "post_payload", "get_clusters", "show",
"get_pubmed", "show#envRefClass", "remove_homologous_interactions",
"get_png"
Reference Superclasses:
"envRefClass"To see a list of methods associated with the STRINGdb class, you can use:
# List all available methods
STRINGdb$methods() [1] ".objectPackage" ".objectParent"
[3] "add_diff_exp_color" "add_proteins_description"
[5] "benchmark_ppi" "benchmark_ppi_pathway_view"
[7] "callSuper" "copy"
[9] "enrichment_heatmap" "export"
[11] "field" "get_aliases"
[13] "get_annotations" "get_bioc_graph"
[15] "get_clusters" "get_enrichment"
[17] "get_graph" "get_homologs"
[19] "get_homologs_besthits" "get_homology_graph"
[21] "get_interactions" "get_link"
[23] "get_neighbors" "get_paralogs"
[25] "get_pathways_benchmarking_blackList" "get_png"
[27] "get_ppi_enrichment" "get_ppi_enrichment_full"
[29] "get_proteins" "get_pubmed"
[31] "get_pubmed_interaction" "get_subnetwork"
[33] "get_summary" "get_term_proteins"
[35] "getClass" "getRefClass"
[37] "import" "initFields"
[39] "initialize" "load"
[41] "load_all" "map"
[43] "mp" "plot_network"
[45] "plot_ppi_enrichment" "post_payload"
[47] "ppi_enrichment" "remove_homologous_interactions"
[49] "set_background" "show"
[51] "show#envRefClass" "trace"
[53] "untrace" "usingMethods" You can also use STRINGdb$help() to access the documentation of each method as follows:
STRINGdb$help("get_graph")Call:
$get_graph()
Description:
Return an igraph object with the entire STRING network.
We invite the user to use the functions of the iGraph package to conveniently
search/analyze the network.
References:
Csardi G, Nepusz T: The igraph software package for complex network research,
InterJournal, Complex Systems 1695. 2006.
http://igraph.sf.net
See Also:
In order to simplify the most common tasks, we do also provide convenient functions
that wrap some iGraph functions.
get_interactions(string_ids) # returns the interactions in between the input proteins
get_neighbors(string_ids) # Get the neighborhoods of a protein (or of a vector of proteins).
get_subnetwork(string_ids) # returns a subgraph from the given input proteins
Author(s):
Andrea FranceschiniOnce we have a connection to STRING for of our species of interest (here, defined in object string_db), we can use the map() method to get STRING protein IDs for our genes in diff_exp_example1.
example1 <- string_db$map(
diff_exp_example1,
"gene",
removeUnmappedRows = TRUE
)Warning: we couldn't map to STRING 15% of your identifiershead(example1) gene pvalue logFC STRING_id
1 VSTM2L 0.0001018 3.333461 9606.ENSP00000362560
2 TBC1D2 0.0001392 3.822383 9606.ENSP00000481721
3 LENG9 0.0001720 3.306056 9606.ENSP00000479355
4 TMEM27 0.0001739 3.024605 9606.ENSP00000369699
5 TSPAN1 0.0002393 3.082052 9606.ENSP00000361072
6 TNNC1 0.0002921 2.932060 9606.ENSP00000232975Next, we can use the plot_network() method to visualize the network for some proteins of interest. Here, let’s use the 200 most significant genes from the differential expression analysis.
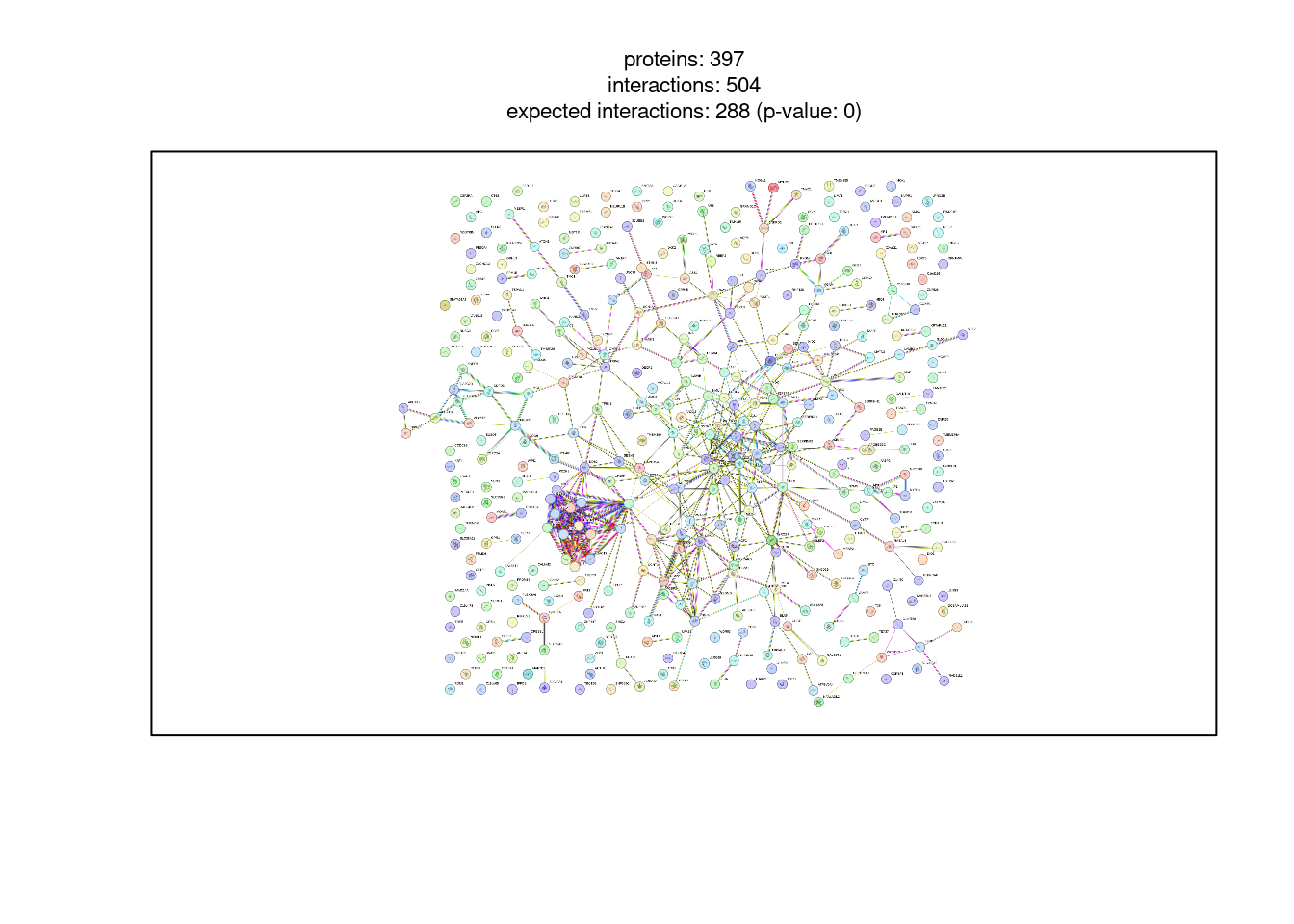
top200 <- example1$STRING_id[1:200]
string_db$plot_network(top200)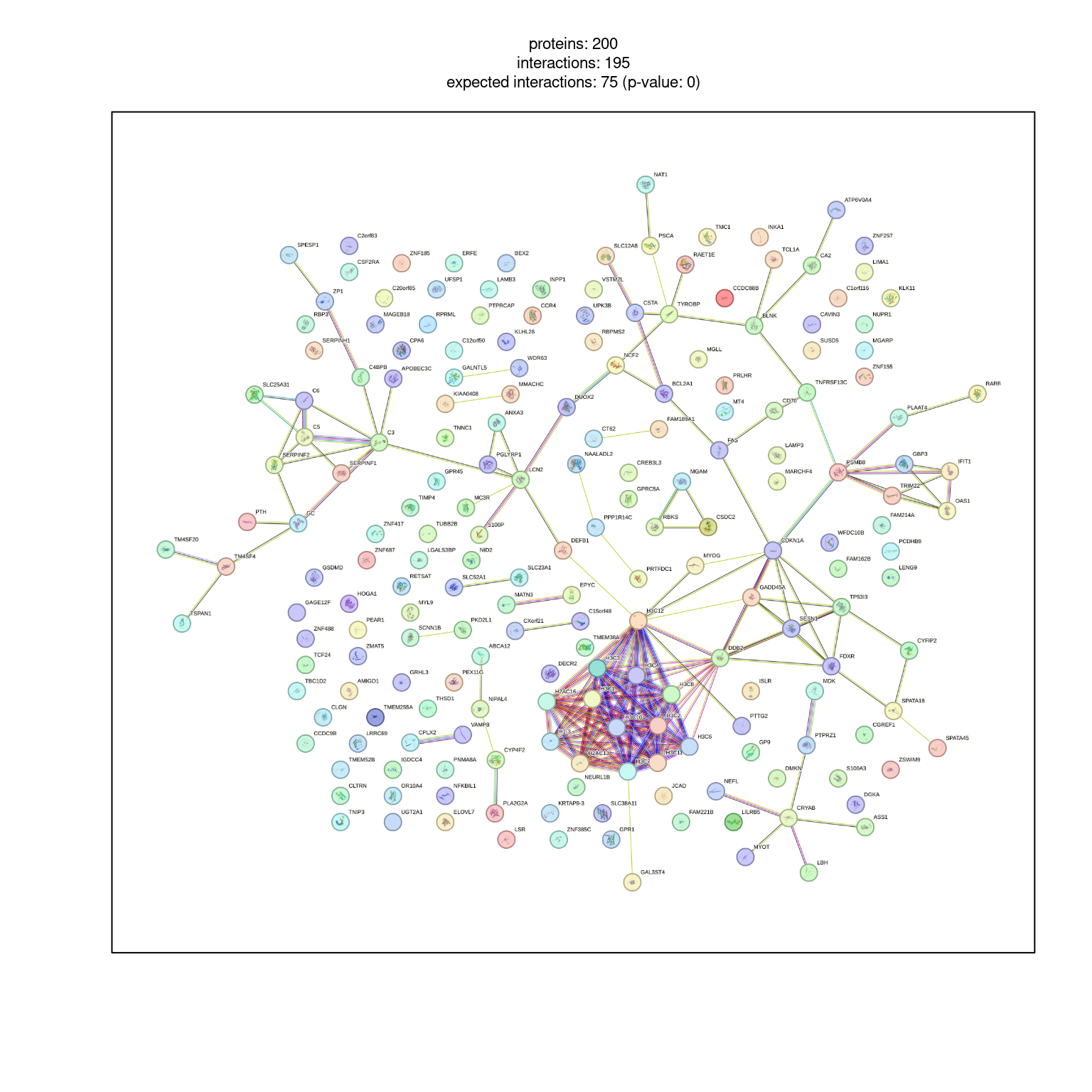
- Plot a PPI network containing only up-regulated genes based on the definition below:
logFC>=1pvalue<0.05.
- Extract the subgraph containing the up-regulated genes identified before (as an object of class
igraph), and then answer the questions below.
- How many nodes and edges are there?
- What are the IDs of the proteins with the largest degrees?
- How many lonely nodes (i.e., nodes with zero connections) are there? What proportion of the total number of nodes does that represent?
4.3 Interacting with the STRING server
STRINGdb allows users to interact with the payload mechanism of STRING, which adds a colored border (or a “halo”) to nodes in visualizations based on some variable. In this sense, we can send data to STRING servers, and use such data to color node borders differently.
To demonstrate this, we will send information on log fold changes, and tell STRING to add colored borders indicating whether genes are up- or down-regulated.
# Remove non-DE genes and add `color` column
example1_filtered <- string_db$add_diff_exp_color(
example1[example1$pvalue < 0.05, ],
logFcColStr = "logFC"
)
head(example1_filtered) gene pvalue logFC STRING_id color
1 VSTM2L 0.0001018 3.333461 9606.ENSP00000362560 #FFDCDCFF
2 TBC1D2 0.0001392 3.822383 9606.ENSP00000481721 #FFC6C6FF
3 LENG9 0.0001720 3.306056 9606.ENSP00000479355 #FFDDDDFF
4 TMEM27 0.0001739 3.024605 9606.ENSP00000369699 #FFE6E6FF
5 TSPAN1 0.0002393 3.082052 9606.ENSP00000361072 #FFE4E4FF
6 TNNC1 0.0002921 2.932060 9606.ENSP00000232975 #FFE8E8FF# Post payload information to STRING server
pid <- string_db$post_payload(
example1_filtered$STRING_id,
colors = example1_filtered$color
)
pid[1] "R_IEKUANQNGJWHA559"# Visualize network with borders ("halo")
string_db$plot_network(top200, payload_id = pid)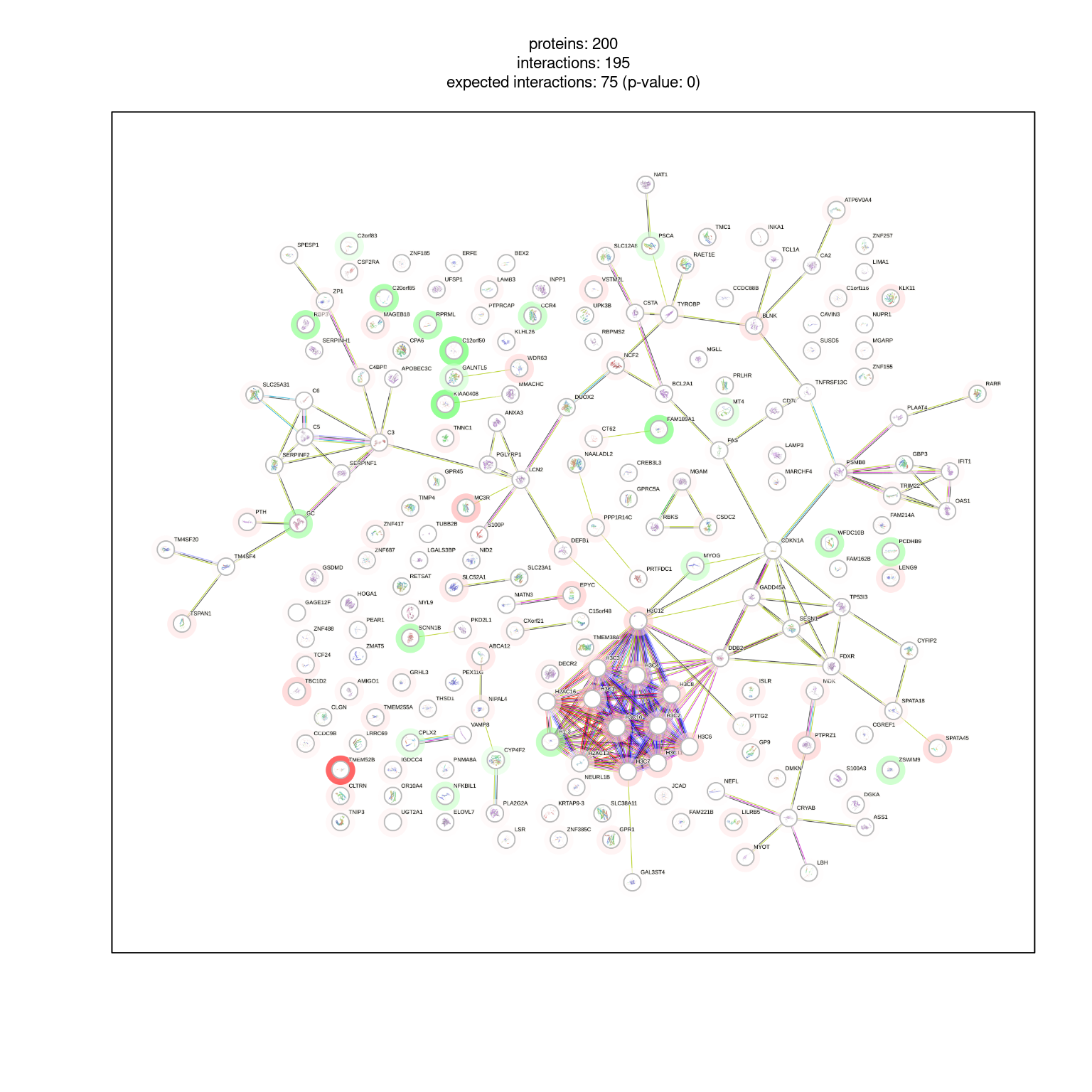
4.4 Overrepresentation analyses
STRINGdb offers a method named get_enrichment() that performs overrepresentation analysis (or “enrichment”) using a hypergeometric test with Benjamini-Hochberg-corrected P-values. For that, the method automatically retrieves data from functional databases, including Gene Ontology, KEGG, Reactome, PFAM, etc.
enrichment <- string_db$get_enrichment(top200)
head(enrichment) category term number_of_genes number_of_genes_in_background
1 COMPARTMENTS GOCC:0000786 10 94
2 COMPARTMENTS GOCC:0032993 12 198
3 COMPARTMENTS GOCC:0005576 42 2079
4 COMPARTMENTS GOCC:0043230 17 524
5 Process GO:0006335 10 32
6 Process GO:0006334 11 124
ncbiTaxonId
1 9606
2 9606
3 9606
4 9606
5 9606
6 9606
inputGenes
1 9606.ENSP00000352252,9606.ENSP00000358160,9606.ENSP00000366999,9606.ENSP00000480826,9606.ENSP00000483283,9606.ENSP00000484095,9606.ENSP00000484638,9606.ENSP00000484658,9606.ENSP00000484841,9606.ENSP00000489282
2 9606.ENSP00000241651,9606.ENSP00000352252,9606.ENSP00000358160,9606.ENSP00000366999,9606.ENSP00000379003,9606.ENSP00000480826,9606.ENSP00000483283,9606.ENSP00000484095,9606.ENSP00000484638,9606.ENSP00000484658,9606.ENSP00000484841,9606.ENSP00000489282
3 9606.ENSP00000008938,9606.ENSP00000216286,9606.ENSP00000223642,9606.ENSP00000243611,9606.ENSP00000245907,9606.ENSP00000254722,9606.ENSP00000258613,9606.ENSP00000259818,9606.ENSP00000262776,9606.ENSP00000263413,9606.ENSP00000264474,9606.ENSP00000264908,9606.ENSP00000278853,9606.ENSP00000285379,9606.ENSP00000287814,9606.ENSP00000297439,9606.ENSP00000301258,9606.ENSP00000321853,9606.ENSP00000345751,9606.ENSP00000349709,9606.ENSP00000352252,9606.ENSP00000361072,9606.ENSP00000362108,9606.ENSP00000362560,9606.ENSP00000371348,9606.ENSP00000373691,9606.ENSP00000375778,9606.ENSP00000377047,9606.ENSP00000379003,9606.ENSP00000383364,9606.ENSP00000383894,9606.ENSP00000385451,9606.ENSP00000395294,9606.ENSP00000421725,9606.ENSP00000433208,9606.ENSP00000433209,9606.ENSP00000442304,9606.ENSP00000447378,9606.ENSP00000463151,9606.ENSP00000473047,9606.ENSP00000480821,9606.ENSP00000498466
4 9606.ENSP00000223642,9606.ENSP00000245907,9606.ENSP00000254722,9606.ENSP00000262776,9606.ENSP00000264908,9606.ENSP00000297439,9606.ENSP00000321853,9606.ENSP00000345751,9606.ENSP00000352252,9606.ENSP00000361072,9606.ENSP00000362108,9606.ENSP00000373691,9606.ENSP00000383364,9606.ENSP00000395294,9606.ENSP00000421725,9606.ENSP00000433209,9606.ENSP00000498466
5 9606.ENSP00000352252,9606.ENSP00000358160,9606.ENSP00000366999,9606.ENSP00000480826,9606.ENSP00000483283,9606.ENSP00000484095,9606.ENSP00000484638,9606.ENSP00000484658,9606.ENSP00000484841,9606.ENSP00000489282
6 9606.ENSP00000244534,9606.ENSP00000352252,9606.ENSP00000358160,9606.ENSP00000366999,9606.ENSP00000480826,9606.ENSP00000483283,9606.ENSP00000484095,9606.ENSP00000484638,9606.ENSP00000484658,9606.ENSP00000484841,9606.ENSP00000489282
preferredNames
1 H3C12,H3C10,H3C4,H3C1,H3C11,H3C7,H3C8,H3C3,H3C2,H3C6
2 MYOG,H3C12,H3C10,H3C4,NUPR1,H3C1,H3C11,H3C7,H3C8,H3C3,H3C2,H3C6
3 PGLYRP1,NID2,C5,C4BPB,C3,SERPINF1,THSD1,TUBB2B,LGALS3BP,C6,CSTA,ANXA3,ZP1,CA2,TIMP4,DEFB1,PSCA,SERPINF2,SCNN1B,RAET1E,H3C12,TSPAN1,LCN2,VSTM2L,TMEM52B,DUOX2,LAMB3,PTPRZ1,NUPR1,PLA2G2A,MATN3,MDK,CD70,GC,PTH,GSDMD,ERFE,MGAM,RBP3,KLK11,LSR,FAS
4 C5,C3,SERPINF1,LGALS3BP,ANXA3,DEFB1,SERPINF2,SCNN1B,H3C12,TSPAN1,LCN2,DUOX2,PLA2G2A,CD70,GC,GSDMD,FAS
5 H3C12,H3C10,H3C4,H3C1,H3C11,H3C7,H3C8,H3C3,H3C2,H3C6
6 H1-3,H3C12,H3C10,H3C4,H3C1,H3C11,H3C7,H3C8,H3C3,H3C2,H3C6
p_value fdr description
1 9.65e-08 2.20e-04 Nucleosome
2 1.50e-06 1.70e-03 protein-DNA complex
3 1.28e-05 7.30e-03 Extracellular region
4 3.51e-05 1.61e-02 Extracellular organelle
5 9.34e-12 1.46e-07 DNA replication-dependent chromatin assembly
6 1.21e-07 6.30e-04 Nucleosome assemblyTo change the background (or “universe”) in your overrepresentation analysis (e.g., to include only expressed genes), you can use the set_background() method. For example:
# Using only genes in `example1` as background
bg <- unique(example1$STRING_id)
string_db$set_background(bg)IGRAPH 5e453a4 UN-- 17501 1719356 --
+ attr: name (v/c), combined_score (e/n)
+ edges from 5e453a4 (vertex names):
[1] 9606.ENSP00000000233--9606.ENSP00000493357
[2] 9606.ENSP00000000233--9606.ENSP00000371175
[3] 9606.ENSP00000000233--9606.ENSP00000354878
[4] 9606.ENSP00000000233--9606.ENSP00000310226
[5] 9606.ENSP00000000233--9606.ENSP00000258098
[6] 9606.ENSP00000000233--9606.ENSP00000363232
[7] 9606.ENSP00000000233--9606.ENSP00000311449
[8] 9606.ENSP00000000233--9606.ENSP00000262455
+ ... omitted several edgesIf, instead of performing overrepresentation analysis, you simply want to obtain functional annotation for your set of proteins, you can use the get_annotations() method as follows.
# Get functional annotation
annot <- string_db$get_annotations(top200)
head(annot) category term_id number_of_genes ratio_in_set species
1 COMPARTMENTS GOCC:0000109 1 0.005 9606
2 COMPARTMENTS GOCC:0000139 1 0.005 9606
3 COMPARTMENTS GOCC:0000151 2 0.010 9606
4 COMPARTMENTS GOCC:0000152 1 0.005 9606
5 COMPARTMENTS GOCC:0000228 10 0.050 9606
6 COMPARTMENTS GOCC:0000307 1 0.005 9606
string_ids
1 9606.ENSP00000256996
2 9606.ENSP00000272895
3 9606.ENSP00000256996,9606.ENSP00000424261
4 9606.ENSP00000424261
5 9606.ENSP00000352252,9606.ENSP00000358160,9606.ENSP00000366999,9606.ENSP00000480826,9606.ENSP00000483283,9606.ENSP00000484095,9606.ENSP00000484638,9606.ENSP00000484658,9606.ENSP00000484841,9606.ENSP00000489282
6 9606.ENSP00000384849
preferred_names
1 DDB2
2 ABCA12
3 DDB2,PTTG2
4 PTTG2
5 H3C12,H3C10,H3C4,H3C1,H3C11,H3C7,H3C8,H3C3,H3C2,H3C6
6 CDKN1A
description
1 Nucleotide-excision repair complex
2 Golgi membrane
3 Ubiquitin ligase complex
4 Nuclear ubiquitin ligase complex
5 Nuclear chromosome
6 Cyclin-dependent protein kinase holoenzyme complexExplore the output of get_annotations() created above and answer the following questions:
- How many proteins are associated with term “Cancer” (DOID:162)?
- What biological processes are associated with gene ZNF155?
4.5 Find PPI clusters
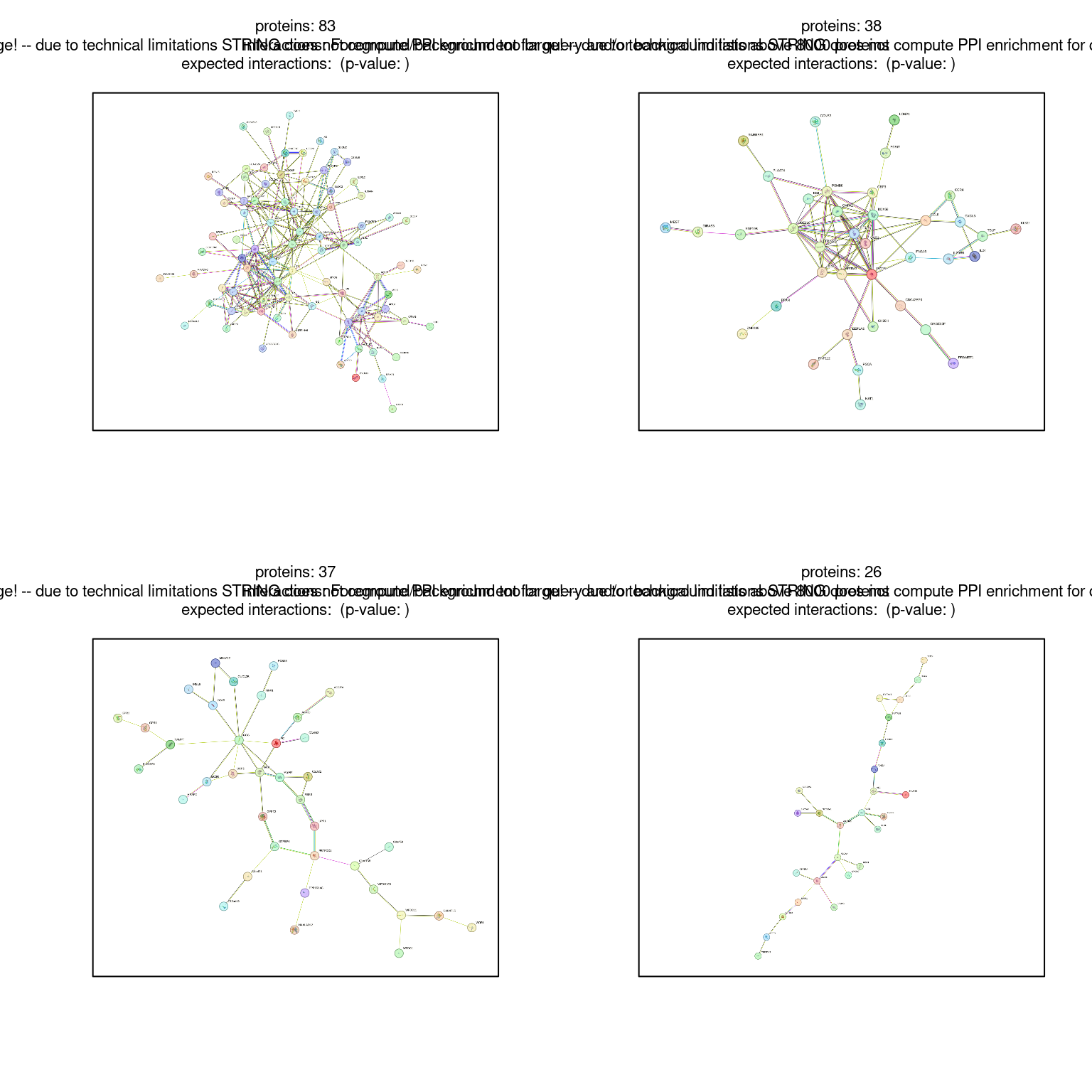
To detect clusters in your PPI network, you can use the get_clusters() method, which in turn uses the clustering algorithms implemented in the cluster_ family of functions in igraph (see ??igraph::cluster_ for a complete list). For example:
# Find clusters using only the first 600 genes
clusters <- string_db$get_clusters(example1$STRING_id[1:600])
# Check first 4 clusters
clusters[1:4][[1]]
[1] "9606.ENSP00000008938" "9606.ENSP00000220166" "9606.ENSP00000220809"
[4] "9606.ENSP00000223642" "9606.ENSP00000225831" "9606.ENSP00000227880"
[7] "9606.ENSP00000228434" "9606.ENSP00000228841" "9606.ENSP00000232975"
[10] "9606.ENSP00000233997" "9606.ENSP00000234347" "9606.ENSP00000239926"
[13] "9606.ENSP00000241651" "9606.ENSP00000243440" "9606.ENSP00000243611"
[16] "9606.ENSP00000244336" "9606.ENSP00000245817" "9606.ENSP00000245907"
[19] "9606.ENSP00000247470" "9606.ENSP00000250092" "9606.ENSP00000254722"
[22] "9606.ENSP00000259212" "9606.ENSP00000259818" "9606.ENSP00000262629"
[25] "9606.ENSP00000263413" "9606.ENSP00000263867" "9606.ENSP00000264192"
[28] "9606.ENSP00000264257" "9606.ENSP00000264474" "9606.ENSP00000264908"
[31] "9606.ENSP00000267953" "9606.ENSP00000279022" "9606.ENSP00000280357"
[34] "9606.ENSP00000281154" "9606.ENSP00000284818" "9606.ENSP00000290378"
[37] "9606.ENSP00000296370" "9606.ENSP00000303942" "9606.ENSP00000308541"
[40] "9606.ENSP00000310036" "9606.ENSP00000321853" "9606.ENSP00000329292"
[43] "9606.ENSP00000329647" "9606.ENSP00000334145" "9606.ENSP00000335397"
[46] "9606.ENSP00000339587" "9606.ENSP00000342012" "9606.ENSP00000342850"
[49] "9606.ENSP00000349709" "9606.ENSP00000352835" "9606.ENSP00000355340"
[52] "9606.ENSP00000356320" "9606.ENSP00000356505" "9606.ENSP00000356906"
[55] "9606.ENSP00000357055" "9606.ENSP00000357981" "9606.ENSP00000358532"
[58] "9606.ENSP00000361850" "9606.ENSP00000362108" "9606.ENSP00000363330"
[61] "9606.ENSP00000363557" "9606.ENSP00000363976" "9606.ENSP00000364555"
[64] "9606.ENSP00000366307" "9606.ENSP00000367817" "9606.ENSP00000367879"
[67] "9606.ENSP00000368341" "9606.ENSP00000373691" "9606.ENSP00000378332"
[70] "9606.ENSP00000378733" "9606.ENSP00000383023" "9606.ENSP00000383840"
[73] "9606.ENSP00000385035" "9606.ENSP00000395294" "9606.ENSP00000404570"
[76] "9606.ENSP00000421725" "9606.ENSP00000433209" "9606.ENSP00000433560"
[79] "9606.ENSP00000435150" "9606.ENSP00000444271" "9606.ENSP00000481699"
[82] "9606.ENSP00000498227" "9606.ENSP00000498466"
[[2]]
[1] "9606.ENSP00000217182" "9606.ENSP00000223215" "9606.ENSP00000237696"
[4] "9606.ENSP00000243346" "9606.ENSP00000253107" "9606.ENSP00000255688"
[7] "9606.ENSP00000256722" "9606.ENSP00000270517" "9606.ENSP00000287156"
[10] "9606.ENSP00000296026" "9606.ENSP00000301258" "9606.ENSP00000328133"
[13] "9606.ENSP00000332134" "9606.ENSP00000332659" "9606.ENSP00000339804"
[16] "9606.ENSP00000353826" "9606.ENSP00000357113" "9606.ENSP00000359512"
[19] "9606.ENSP00000360020" "9606.ENSP00000360869" "9606.ENSP00000360918"
[22] "9606.ENSP00000362555" "9606.ENSP00000364016" "9606.ENSP00000369213"
[25] "9606.ENSP00000369299" "9606.ENSP00000369564" "9606.ENSP00000375795"
[28] "9606.ENSP00000378118" "9606.ENSP00000382707" "9606.ENSP00000388001"
[31] "9606.ENSP00000429089" "9606.ENSP00000443194" "9606.ENSP00000454919"
[34] "9606.ENSP00000464788" "9606.ENSP00000473047" "9606.ENSP00000480549"
[37] "9606.ENSP00000491865" "9606.ENSP00000496832"
[[3]]
[1] "9606.ENSP00000016913" "9606.ENSP00000222553" "9606.ENSP00000223366"
[4] "9606.ENSP00000243911" "9606.ENSP00000252971" "9606.ENSP00000253778"
[7] "9606.ENSP00000257776" "9606.ENSP00000294664" "9606.ENSP00000302485"
[10] "9606.ENSP00000302724" "9606.ENSP00000306817" "9606.ENSP00000307292"
[13] "9606.ENSP00000316598" "9606.ENSP00000318602" "9606.ENSP00000323071"
[16] "9606.ENSP00000328938" "9606.ENSP00000330032" "9606.ENSP00000337466"
[19] "9606.ENSP00000341610" "9606.ENSP00000348968" "9606.ENSP00000355260"
[22] "9606.ENSP00000361662" "9606.ENSP00000363136" "9606.ENSP00000368245"
[25] "9606.ENSP00000370997" "9606.ENSP00000371054" "9606.ENSP00000377112"
[28] "9606.ENSP00000387662" "9606.ENSP00000404705" "9606.ENSP00000446280"
[31] "9606.ENSP00000447378" "9606.ENSP00000456920" "9606.ENSP00000477802"
[34] "9606.ENSP00000479207" "9606.ENSP00000480405" "9606.ENSP00000498994"
[37] "9606.ENSP00000499695"
[[4]]
[1] "9606.ENSP00000075120" "9606.ENSP00000078445" "9606.ENSP00000220966"
[4] "9606.ENSP00000223836" "9606.ENSP00000265598" "9606.ENSP00000295213"
[7] "9606.ENSP00000301732" "9606.ENSP00000302851" "9606.ENSP00000307142"
[10] "9606.ENSP00000310658" "9606.ENSP00000327213" "9606.ENSP00000334910"
[13] "9606.ENSP00000357811" "9606.ENSP00000359245" "9606.ENSP00000361471"
[16] "9606.ENSP00000370737" "9606.ENSP00000377176" "9606.ENSP00000388895"
[19] "9606.ENSP00000394227" "9606.ENSP00000399979" "9606.ENSP00000417161"
[22] "9606.ENSP00000419160" "9606.ENSP00000427679" "9606.ENSP00000463151"
[25] "9606.ENSP00000477707" "9606.ENSP00000479719"# Plotting first 4 clusters
par(mfrow = c(2,2))
for(x in seq_len(4)) {
string_db$plot_network(clusters[[x]])
}
par(mfrow = c(1,1)) # back to original number of rows and cols4.6 Extracting extra information for proteins of interest
Besides obtaining STRING protein IDs from a pre-defined list of IDs, you can also extract all proteins in the STRING database (for the organism you specified when creating the connection). This can be achiveved with the get_proteins() method.
# Get a list of all proteins
all_proteins <- string_db$get_proteins()
head(all_proteins) protein_external_id preferred_name protein_size
1 9606.ENSP00000000233 ARF5 180
2 9606.ENSP00000000412 M6PR 277
3 9606.ENSP00000001008 FKBP4 459
4 9606.ENSP00000001146 CYP26B1 512
5 9606.ENSP00000002125 NDUFAF7 441
6 9606.ENSP00000002165 FUCA2 467
annotation
1 ADP-ribosylation factor 5; GTP-binding protein involved in protein trafficking; may modulate vesicle budding and uncoating within the Golgi apparatus. Belongs to the small GTPase superfamily. Arf family.
2 Cation-dependent mannose-6-phosphate receptor; Transport of phosphorylated lysosomal enzymes from the Golgi complex and the cell surface to lysosomes. Lysosomal enzymes bearing phosphomannosyl residues bind specifically to mannose-6-phosphate receptors in the Golgi apparatus and the resulting receptor-ligand complex is transported to an acidic prelyosomal compartment where the low pH mediates the dissociation of the complex.
3 Peptidyl-prolyl cis-trans isomerase FKBP4, N-terminally processed; Immunophilin protein with PPIase and co-chaperone activities. Component of steroid receptors heterocomplexes through interaction with heat-shock protein 90 (HSP90). May play a role in the intracellular trafficking of heterooligomeric forms of steroid hormone receptors between cytoplasm and nuclear compartments. The isomerase activity controls neuronal growth cones via regulation of TRPC1 channel opening. Acts also as a regulator of microtubule dynamics by inhibiting MAPT/TAU ability to promote microtubule assembly. May [...]
4 Cytochrome P450 26B1; Involved in the metabolism of retinoic acid (RA), rendering this classical morphogen inactive through oxidation. Involved in the specific inactivation of all-trans-retinoic acid (all-trans-RA), with a preference for the following substrates: all-trans-RA > 9-cis-RA > 13- cis-RA. Generates several hydroxylated forms of RA, including 4-OH-RA, 4-oxo-RA, and 18-OH-RA. Catalyzes the hydroxylation of carbon hydrogen bonds of atRA primarily at C-4. Essential for postnatal survival. Plays a central role in germ cell development: acts by degrading RA in the developing test [...]
5 Protein arginine methyltransferase NDUFAF7, mitochondrial; Arginine methyltransferase involved in the assembly or stability of mitochondrial NADH:ubiquinone oxidoreductase complex (complex I). Acts by mediating symmetric dimethylation of 'Arg-118' of NDUFS2 after it assembles into the complex I, stabilizing the early intermediate complex.
6 Plasma alpha-L-fucosidase; Alpha-L-fucosidase is responsible for hydrolyzing the alpha- 1,6-linked fucose joined to the reducing-end N-acetylglucosamine of the carbohydrate moieties of glycoproteins; Belongs to the glycosyl hydrolase 29 family.If you want only the STRING IDs (as a vector) from an input vector of protein IDs, you can use the mp() method. This works very similarly to the map() method we used before, but this one returns a vector, not a data frame.
# Get STRING ID for proteins 'tp53' and 'atm'
tp53 <- string_db$mp("tp53")
atm <- string_db$mp("atm")
list(tp53, atm)[[1]]
[1] "9606.ENSP00000269305"
[[2]]
[1] "9606.ENSP00000278616"You can also retrieve all proteins that interact with a protein of interest (i.e., the neighbors of a protein of interest) using the get_neighbors() method.
# Get neighbors of 'tp53'
string_db$get_neighbors(tp53) |> head()[1] "9606.ENSP00000001008" "9606.ENSP00000003084" "9606.ENSP00000003302"
[4] "9606.ENSP00000004982" "9606.ENSP00000005257" "9606.ENSP00000005260"We can also use the get_interactions() method to get interactions between input proteins (if any). For example, to check if proteins TP53 and ATM interact, we can use:
# Get connections between tp53 and ATM
string_db$get_interactions(c(tp53, atm)) from to combined_score
1 9606.ENSP00000269305 9606.ENSP00000278616 999
2 9606.ENSP00000269305 9606.ENSP00000278616 999Which protein has more interaction partners: TP53 or ATM?
Are there proteins that interact with both TP53 and ATM? If so, how many?
4.7 Extracting homologous proteins
Finally, STRINGdb can also be used to extract homologous proteins (paralogs and/or orthologs) of proteins of interest. To extract all paralogs of a specific protein, you will use the get_paralogs() method as follows:
# Extract paralogs of tp53 (if any)
string_db$get_paralogs(tp53)[1] X9606 X9606.ENSP00000269305 X9606.1
[4] X9606.ENSP00000269305.1 X815.8
<0 rows> (or 0-length row.names)string_db$get_homologs_besthits(tp53) X9606 X9606.ENSP00000269305 X9597 X9597.ENSPPAP00000011040 X815.8
1 9606 9606.ENSP00000269305 9598 9598.ENSPTRP00000014836 815.8
2 9606 9606.ENSP00000269305 9606 9606.ENSP00000269305 815.8
3 9606 9606.ENSP00000269305 9601 9601.ENSPPYP00000008923 799.3
4 9606 9606.ENSP00000269305 61621 61621.ENSRBIP00000028632 786.6
5 9606 9606.ENSP00000269305 61622 61622.ENSRROP00000026843 786.6
6 9606 9606.ENSP00000269305 591936 591936.ENSPTEP00000010553 785.8
7 9606 9606.ENSP00000269305 60711 60711.ENSCSAP00000007551 785.0
8 9606 9606.ENSP00000269305 9541 9541.ENSMFAP00000038526 783.9
9 9606 9606.ENSP00000269305 9544 9544.ENSMMUP00000011334 783.9
10 9606 9606.ENSP00000269305 9565 9565.ENSTGEP00000031378 783.1
11 9606 9606.ENSP00000269305 9531 9531.ENSCATP00000032045 738.8
12 9606 9606.ENSP00000269305 9568 9568.ENSMLEP00000037099 738.4
13 9606 9606.ENSP00000269305 9555 9555.ENSPANP00000033477 737.3
14 9606 9606.ENSP00000269305 336983 336983.ENSCANP00000028904 735.7
15 9606 9606.ENSP00000269305 9516 9516.ENSCCAP00000029525 729.6
16 9606 9606.ENSP00000269305 1868482 1868482.ENSTSYP00000023752 707.6
17 9606 9606.ENSP00000269305 379532 379532.ENSPCOP00000031758 704.1
18 9606 9606.ENSP00000269305 61853 61853.ENSNLEP00000011861 702.6
19 9606 9606.ENSP00000269305 9994 9994.ENSMMMP00000022491 699.1
20 9606 9606.ENSP00000269305 43179 43179.ENSSTOP00000019678 697.2
21 9606 9606.ENSP00000269305 9999 9999.ENSUPAP00010019485 696.4
22 9606 9606.ENSP00000269305 99837 99837.ENSSDAP00000002829 696.0
23 9606 9606.ENSP00000269305 9986 9986.ENSOCUP00000000989 683.7
24 9606 9606.ENSP00000269305 9483 9483.ENSCJAP00000066800 682.6
25 9606 9606.ENSP00000269305 30608 30608.ENSMICP00000039812 681.8
26 9606 9606.ENSP00000269305 1026970 1026970.ENSNGAP00000007306 679.9
27 9606 9606.ENSP00000269305 9755 9755.ENSPCTP00005020186 676.8
28 9606 9606.ENSP00000269305 9749 9749.Q8SPZ3 676.4
29 9606 9606.ENSP00000269305 118797 118797.A0A340XCN5 674.9
30 9606 9606.ENSP00000269305 1706337 1706337.A0A341BQX3 671.4
31 9606 9606.ENSP00000269305 9593 9593.ENSGGOP00000030954 671.4
32 9606 9606.ENSP00000269305 310752 310752.A0A383YUA7 669.8
33 9606 9606.ENSP00000269305 9739 9739.ENSTTRP00000009721 668.3
34 9606 9606.ENSP00000269305 10181 10181.G5B5D6 662.1
35 9606 9606.ENSP00000269305 9838 9838.ENSCDRP00005013165 656.8
36 9606 9606.ENSP00000269305 39432 39432.ENSSBOP00000008181 656.4
37 9606 9606.ENSP00000269305 30611 30611.ENSOGAP00000015106 655.6
38 9606 9606.ENSP00000269305 34839 34839.ENSCLAP00000010404 655.6
39 9606 9606.ENSP00000269305 9796 9796.ENSECAP00000032751 655.2
40 9606 9606.ENSP00000269305 83772 83772.ENSEASP00005028399 653.7
41 9606 9606.ENSP00000269305 51154 51154.ENSCWAP00000005800 652.9
42 9606 9606.ENSP00000269305 230844 230844.ENSPEMP00000010300 651.7
43 9606 9606.ENSP00000269305 68415 68415.ENSMMSP00000026578 640.6
44 9606 9606.ENSP00000269305 9646 9646.ENSAMEP00000017825 640.2
45 9606 9606.ENSP00000269305 885580 885580.ENSFDAP00000011613 636.3
46 9606 9606.ENSP00000269305 9643 9643.ENSUAMP00000000913 635.6
47 9606 9606.ENSP00000269305 116960 116960.A0A3Q7Y5W2 634.4
48 9606 9606.ENSP00000269305 29073 29073.ENSUMAP00000023207 634.4
49 9606 9606.ENSP00000269305 286419 286419.ENSCAFP00020009510 634.0
50 9606 9606.ENSP00000269305 9708 9708.A0A2U3VT95 634.0
51 9606 9606.ENSP00000269305 9361 9361.ENSDNOP00000004075 633.6
52 9606 9606.ENSP00000269305 9615 9615.ENSCAFP00000064590 633.3
53 9606 9606.ENSP00000269305 9691 9691.ENSPPRP00000022844 633.3
54 9606 9606.ENSP00000269305 59479 59479.ENSRFEP00010016808 631.7
55 9606 9606.ENSP00000269305 9627 9627.ENSVVUP00000039187 631.7
56 9606 9606.ENSP00000269305 9713 9713.A0A2U3Y5G3 630.9
57 9606 9606.ENSP00000269305 29088 29088.A0A2Y9HEV7 629.8
58 9606 9606.ENSP00000269305 10160 10160.ENSODEP00000024162 624.8
59 9606 9606.ENSP00000269305 72004 72004.ENSBMUP00000028863 622.9
60 9606 9606.ENSP00000269305 30521 30521.ENSBGRP00000021669 622.9
61 9606 9606.ENSP00000269305 43346 43346.ENSBBBP00000018891 622.9
62 9606 9606.ENSP00000269305 79684 79684.ENSMOCP00000023219 621.3
63 9606 9606.ENSP00000269305 391180 391180.A0A2Y9L4Q2 620.5
64 9606 9606.ENSP00000269305 9913 9913.ENSBTAP00000001420 620.2
65 9606 9606.ENSP00000269305 452646 452646.ENSNVIP00000005141 619.4
66 9606 9606.ENSP00000269305 9925 9925.ENSCHIP00000030141 618.2
67 9606 9606.ENSP00000269305 9669 9669.ENSMPUP00000008945 617.8
68 9606 9606.ENSP00000269305 30522 30522.A0A4W2I2P0 617.5
69 9606 9606.ENSP00000269305 42254 42254.ENSSARP00000005616 617.1
70 9606 9606.ENSP00000269305 51337 51337.ENSJJAP00000008090 616.7
71 9606 9606.ENSP00000269305 9940 9940.ENSOARP00000016707 616.7
72 9606 9606.ENSP00000269305 37032 37032.ENSSSUP00005028514 616.7
73 9606 9606.ENSP00000269305 61383 61383.ENSLCNP00005018106 615.5
74 9606 9606.ENSP00000269305 127582 127582.A0A2Y9R5B9 613.6
75 9606 9606.ENSP00000269305 9685 9685.ENSFCAP00000045749 612.1
76 9606 9606.ENSP00000269305 10089 10089.MGP_CAROLIEiJ_P0027554 606.3
77 9606 9606.ENSP00000269305 51338 51338.ENSCCNP00000017223 604.7
78 9606 9606.ENSP00000269305 10096 10096.MGP_SPRETEiJ_P0028933 603.6
79 9606 9606.ENSP00000269305 10093 10093.MGP_PahariEiJ_P0034306 601.3
80 9606 9606.ENSP00000269305 9402 9402.L5JZ91 600.1
81 9606 9606.ENSP00000269305 10116 10116.ENSRNOP00000074031 600.1
82 9606 9606.ENSP00000269305 37548 37548.ENSCAPP00000017521 600.1
83 9606 9606.ENSP00000269305 10141 10141.ENSCPOP00000029751 599.0
84 9606 9606.ENSP00000269305 10090 10090.ENSMUSP00000104298 598.2
85 9606 9606.ENSP00000269305 10036 10036.ENSMAUP00000025852 597.8
86 9606 9606.ENSP00000269305 1328070 1328070.ENSPSMP00000026875 595.5
87 9606 9606.ENSP00000269305 10103 10103.ENSMSIP00000031034 595.1
88 9606 9606.ENSP00000269305 10047 10047.ENSMUGP00000017526 593.6
89 9606 9606.ENSP00000269305 9785 9785.ENSLAFP00000006292 592.0
90 9606 9606.ENSP00000269305 9823 9823.ENSSSCP00000050664 591.7
91 9606 9606.ENSP00000269305 9978 9978.ENSOPRP00000005906 586.3
92 9606 9606.ENSP00000269305 246437 246437.L9KM90 574.3
93 9606 9606.ENSP00000269305 9813 9813.ENSPCAP00000011892 568.5
94 9606 9606.ENSP00000269305 10020 10020.ENSDORP00000023664 562.4
95 9606 9606.ENSP00000269305 59463 59463.ENSMLUP00000005861 559.3
96 9606 9606.ENSP00000269305 37293 37293.ENSANAP00000025592 547.4
97 9606 9606.ENSP00000269305 38626 38626.ENSPCIP00000039513 502.3
98 9606 9606.ENSP00000269305 29139 29139.ENSVURP00010004263 496.1
99 9606 9606.ENSP00000269305 9305 9305.ENSSHAP00000018268 486.1
100 9606 9606.ENSP00000269305 9365 9365.ENSEEUP00000013295 477.6
101 9606 9606.ENSP00000269305 9371 9371.ENSETEP00000007296 420.2
102 9606 9606.ENSP00000269305 132908 132908.ENSPVAP00000015400 415.6
103 9606 9606.ENSP00000269305 9545 9545.ENSMNEP00000030865 399.8
104 9606 9606.ENSP00000269305 56216 56216.A0A1A6GWM4 394.0
105 9606 9606.ENSP00000269305 7897 7897.ENSLACP00000015915 393.3
106 9606 9606.ENSP00000269305 8496 8496.A0A151MW63 373.6
107 9606 9606.ENSP00000269305 2587831 2587831.ENSTMTP00000028378 372.5
108 9606 9606.ENSP00000269305 9315 9315.ENSMEUP00000011978 371.7
109 9606 9606.ENSP00000269305 8478 8478.ENSCPBP00000031582 364.8
110 9606 9606.ENSP00000269305 113540 113540.ENSSFOP00015035795 361.3
111 9606 9606.ENSP00000269305 38654 38654.A0A3Q0FUE5 359.8
112 9606 9606.ENSP00000269305 38772 38772.ENSGAGP00000006237 359.8
113 9606 9606.ENSP00000269305 13735 13735.ENSPSIP00000014905 359.4
114 9606 9606.ENSP00000269305 106734 106734.ENSCABP00000018274 359.0
115 9606 9606.ENSP00000269305 7918 7918.ENSLOCP00000017063 359.0
116 9606 9606.ENSP00000269305 62062 62062.ENSHHUP00000013468 357.1
117 9606 9606.ENSP00000269305 1676925 1676925.ENSPKIP00000014805 357.1
118 9606 9606.ENSP00000269305 8032 8032.ENSSTUP00000101309 356.7
119 9606 9606.ENSP00000269305 75366 75366.ENSSGRP00000074183 356.3
120 9606 9606.ENSP00000269305 8030 8030.ENSSSAP00000057928 355.5
121 9606 9606.ENSP00000269305 307959 307959.ENSSRHP00000051429 353.2
122 9606 9606.ENSP00000269305 1608454 1608454.ENSSANP00000026698 352.8
123 9606 9606.ENSP00000269305 27687 27687.ENSECRP00000015713 351.7
124 9606 9606.ENSP00000269305 8005 8005.ENSEEEP00000033982 345.9
125 9606 9606.ENSP00000269305 8010 8010.ENSELUP00000026168 344.0
126 9606 9606.ENSP00000269305 7998 7998.ENSIPUP00000031919 343.6
127 9606 9606.ENSP00000269305 74533 74533.ENSPTIP00000024834 343.2
128 9606 9606.ENSP00000269305 96440 96440.ENSSMRP00000003871 340.9
129 9606 9606.ENSP00000269305 7955 7955.ENSDARP00000116736 340.9
130 9606 9606.ENSP00000269305 84645 84645.A0A498NKF5 340.1
131 9606 9606.ENSP00000269305 8364 8364.ENSXETP00000041856 339.7
132 9606 9606.ENSP00000269305 37347 37347.ENSTBEP00000010189 336.7
133 9606 9606.ENSP00000269305 299321 299321.ENSDCDP00000007017 332.0
134 9606 9606.ENSP00000269305 8469 8469.M7CJF0 330.9
135 9606 9606.ENSP00000269305 103695 103695.ENSPVIP00000017506 324.3
136 9606 9606.ENSP00000269305 623744 623744.A0A553PV69 319.7
137 9606 9606.ENSP00000269305 8508 8508.ENSSPUP00000018842 313.9
138 9606 9606.ENSP00000269305 2489341 2489341.ENSSHBP00005007544 313.2
139 9606 9606.ENSP00000269305 42514 42514.ENSPNAP00000014501 313.2
140 9606 9606.ENSP00000269305 61221 61221.ENSVKKP00000029045 310.8
141 9606 9606.ENSP00000269305 7994 7994.ENSAMXP00000021366 309.3
142 9606 9606.ENSP00000269305 7950 7950.ENSCHAP00000042143 305.1
143 9606 9606.ENSP00000269305 586833 586833.ENSMMDP00005051289 304.7
144 9606 9606.ENSP00000269305 8022 8022.A0A060XN97 304.3
145 9606 9606.ENSP00000269305 8081 8081.ENSPREP00000019676 293.5
146 9606 9606.ENSP00000269305 52904 52904.ENSSMAP00000015064 292.0
147 9606 9606.ENSP00000269305 47969 47969.ENSOABP00000003944 291.6
148 9606 9606.ENSP00000269305 181472 181472.ENSSFAP00005012771 291.6
149 9606 9606.ENSP00000269305 375764 375764.ENSSORP00005058467 291.6
150 9606 9606.ENSP00000269305 8153 8153.ENSHBUP00000022213 291.2
151 9606 9606.ENSP00000269305 8167 8167.A0A484CCM0 291.2
152 9606 9606.ENSP00000269305 215358 215358.ENSLCRP00005011976 290.8
153 9606 9606.ENSP00000269305 80972 80972.ENSAOCP00000001385 290.0
154 9606 9606.ENSP00000269305 161767 161767.ENSAPEP00000017927 290.0
155 9606 9606.ENSP00000269305 8083 8083.ENSXMAP00000028829 290.0
156 9606 9606.ENSP00000269305 37003 37003.ENSKMAP00000018969 289.7
157 9606 9606.ENSP00000269305 64176 64176.ENSPMRP00000017811 289.3
158 9606 9606.ENSP00000269305 441366 441366.ENSGWIP00000038103 288.9
159 9606 9606.ENSP00000269305 8128 8128.ENSONIP00000041809 287.7
160 9606 9606.ENSP00000269305 80966 80966.ENSAPOP00000027715 287.3
161 9606 9606.ENSP00000269305 31033 31033.ENSTRUP00000030644 287.0
162 9606 9606.ENSP00000269305 32473 32473.ENSXCOP00000002276 285.8
163 9606 9606.ENSP00000269305 48701 48701.ENSPMEP00000001243 285.4
164 9606 9606.ENSP00000269305 244447 244447.ENSCSEP00000003997 285.4
165 9606 9606.ENSP00000269305 8154 8154.ENSACLP00000007040 285.0
166 9606 9606.ENSP00000269305 8175 8175.ENSSAUP00010029879 285.0
167 9606 9606.ENSP00000269305 48698 48698.ENSPFOP00000007556 284.6
168 9606 9606.ENSP00000269305 56723 56723.ENSLBEP00000036857 284.6
169 9606 9606.ENSP00000269305 64144 64144.ENSATEP00000028323 284.3
170 9606 9606.ENSP00000269305 8187 8187.ENSLCAP00010016052 283.9
171 9606 9606.ENSP00000269305 210632 210632.ENSPRNP00000030862 283.5
172 9606 9606.ENSP00000269305 48699 48699.ENSPLAP00000022694 283.1
173 9606 9606.ENSP00000269305 61819 61819.ENSACIP00000020373 282.3
174 9606 9606.ENSP00000269305 144197 144197.ENSSPAP00000022019 282.3
175 9606 9606.ENSP00000269305 299123 299123.ENSLSDP00000017092 280.4
176 9606 9606.ENSP00000269305 56716 56716.ENSCGOP00000014430 279.6
177 9606 9606.ENSP00000269305 205130 205130.ENSMAMP00000005315 279.6
178 9606 9606.ENSP00000269305 137246 137246.A0A401SCC1 279.3
179 9606 9606.ENSP00000269305 99883 99883.ENSTNIP00000013035 278.1
180 9606 9606.ENSP00000269305 173247 173247.ENSENLP00000038795 277.3
181 9606 9606.ENSP00000269305 8078 8078.ENSFHEP00000026765 276.6
182 9606 9606.ENSP00000269305 41447 41447.ENSSDUP00000028226 276.6
183 9606 9606.ENSP00000269305 59729 59729.ENSTGUP00000021519 276.2
184 9606 9606.ENSP00000269305 28743 28743.ENSCVAP00000022398 276.2
185 9606 9606.ENSP00000269305 33528 33528.ENSGAFP00000008062 276.2
186 9606 9606.ENSP00000269305 43700 43700.ENSMALP00000000877 275.8
187 9606 9606.ENSP00000269305 47308 47308.ENSNMLP00000021935 275.8
188 9606 9606.ENSP00000269305 143630 143630.A0A402EXU8 275.4
189 9606 9606.ENSP00000269305 240159 240159.A0A4U5UJG8 275.4
190 9606 9606.ENSP00000269305 52670 52670.A0A2I4CET8 275.0
191 9606 9606.ENSP00000269305 69293 69293.ENSGACP00000018012 275.0
192 9606 9606.ENSP00000269305 106582 106582.ENSMZEP00005006261 274.6
193 9606 9606.ENSP00000269305 8049 8049.ENSGMOP00000000401 274.6
194 9606 9606.ENSP00000269305 303518 303518.ENSPNYP00000025442 274.2
195 9606 9606.ENSP00000269305 1841481 1841481.ENSSLDP00000032981 274.2
196 9606 9606.ENSP00000269305 158456 158456.ENSBSLP00000007319 273.9
197 9606 9606.ENSP00000269305 8355 8355.A0A1L8FGI1 273.9
198 9606 9606.ENSP00000269305 28377 28377.ENSACAP00000001746 273.9
199 9606 9606.ENSP00000269305 75743 75743.A0A401P193 273.5
200 9606 9606.ENSP00000269305 109280 109280.ENSHCOP00000022970 273.1
201 9606 9606.ENSP00000269305 8673 8673.ENSPTXP00000024136 273.1
202 9606 9606.ENSP00000269305 8090 8090.ENSORLP00000019616 272.7
203 9606 9606.ENSP00000269305 9089 9089.ENSCPIP00010009060 271.9
204 9606 9606.ENSP00000269305 30732 30732.ENSOMEP00000009309 271.9
205 9606 9606.ENSP00000269305 59894 59894.ENSFALP00000010031 271.6
206 9606 9606.ENSP00000269305 156563 156563.ENSCCEP00000019692 271.6
207 9606 9606.ENSP00000269305 12930 12930.A0A0Q3T504 271.6
208 9606 9606.ENSP00000269305 333673 333673.A0A3M0IUV8 271.2
209 9606 9606.ENSP00000269305 8996 8996.ENSNMEP00000004526 271.2
210 9606 9606.ENSP00000269305 9157 9157.ENSPMJP00000017734 271.2
211 9606 9606.ENSP00000269305 44394 44394.ENSZALP00000002887 270.8
212 9606 9606.ENSP00000269305 9135 9135.ENSSCAP00000001137 270.8
213 9606 9606.ENSP00000269305 93934 93934.ENSCJPP00005010805 270.4
214 9606 9606.ENSP00000269305 409849 409849.ENSPMGP00000017230 270.4
215 9606 9606.ENSP00000269305 9031 9031.ENSGALP00000001485 270.4
216 9606 9606.ENSP00000269305 9049 9049.ENSPSTP00000007061 270.4
217 9606 9606.ENSP00000269305 9054 9054.ENSPCLP00000024156 270.4
218 9606 9606.ENSP00000269305 13146 13146.ENSMUNP00000017973 270.4
219 9606 9606.ENSP00000269305 372326 372326.A0A1V4KNI0 270.0
220 9606 9606.ENSP00000269305 13616 13616.ENSMODP00000018831 270.0
221 9606 9606.ENSP00000269305 94827 94827.A0A099ZQ65 269.6
222 9606 9606.ENSP00000269305 44316 44316.ENSEGOP00005018987 269.2
223 9606 9606.ENSP00000269305 55661 55661.A0A091G407 269.2
224 9606 9606.ENSP00000269305 94237 94237.ENSMMOP00000018694 269.2
225 9606 9606.ENSP00000269305 128390 128390.A0A091UUY0 269.2
226 9606 9606.ENSP00000269305 441894 441894.ENSSCUP00000005331 269.2
227 9606 9606.ENSP00000269305 7868 7868.ENSCMIP00000007753 269.2
228 9606 9606.ENSP00000269305 9238 9238.A0A093R114 269.2
229 9606 9606.ENSP00000269305 55544 55544.A0A4D9E5P9 268.9
230 9606 9606.ENSP00000269305 85066 85066.A0A091EA06 268.9
231 9606 9606.ENSP00000269305 198806 198806.ENSCPUP00000025340 268.9
232 9606 9606.ENSP00000269305 308060 308060.ENSARWP00000009421 268.9
233 9606 9606.ENSP00000269305 425635 425635.ENSCPGP00000026631 268.9
234 9606 9606.ENSP00000269305 8502 8502.ENSCPRP00005017750 268.9
235 9606 9606.ENSP00000269305 8790 8790.ENSDNVP00000009001 268.9
236 9606 9606.ENSP00000269305 8823 8823.ENSAHAP00000009000 268.9
237 9606 9606.ENSP00000269305 8824 8824.ENSAOWP00000012514 268.9
238 9606 9606.ENSP00000269305 9244 9244.A0A091HZ84 268.9
239 9606 9606.ENSP00000269305 30464 30464.ENSNPEP00000010833 268.9
240 9606 9606.ENSP00000269305 40217 40217.ENSJHYP00000001101 268.9
241 9606 9606.ENSP00000269305 50402 50402.A0A0A0AU13 268.5
242 9606 9606.ENSP00000269305 118200 118200.A0A093GLW0 268.5
243 9606 9606.ENSP00000269305 132585 132585.ENSABRP00000009434 268.5
244 9606 9606.ENSP00000269305 194338 194338.ENSACUP00000017269 268.5
245 9606 9606.ENSP00000269305 211598 211598.ENSANIP00000006686 268.5
246 9606 9606.ENSP00000269305 241587 241587.ENSACOP00000010351 268.5
247 9606 9606.ENSP00000269305 8840 8840.ENSAPLP00000032186 268.5
248 9606 9606.ENSP00000269305 8845 8845.ENSACDP00005018035 268.5
249 9606 9606.ENSP00000269305 8932 8932.A0A2I0LU30 268.5
250 9606 9606.ENSP00000269305 30419 30419.A0A091VW97 268.5
251 9606 9606.ENSP00000269305 223781 223781.ENSACCP00020004462 268.1
252 9606 9606.ENSP00000269305 328815 328815.ENSMVIP00005001759 268.1
253 9606 9606.ENSP00000269305 9233 9233.A0A087R7Z2 268.1
254 9606 9606.ENSP00000269305 9258 9258.ENSOANP00000021753 268.1
255 9606 9606.ENSP00000269305 321398 321398.ENSLCOP00000004510 267.7
256 9606 9606.ENSP00000269305 10029 10029.G3GT84 266.9
257 9606 9606.ENSP00000269305 8663 8663.ENSNSUP00000003328 264.6
258 9606 9606.ENSP00000269305 7757 7757.ENSPMAP00000010479 258.1
259 9606 9606.ENSP00000269305 32507 32507.ENSNBRP00000026855 257.7
260 9606 9606.ENSP00000269305 9009 9009.A0A226MN96 255.4
261 9606 9606.ENSP00000269305 7764 7764.ENSEBUP00000013202 251.9
262 9606 9606.ENSP00000269305 9358 9358.ENSCHOP00000009300 241.9
263 9606 9606.ENSP00000269305 7739 7739.C3XPU2 230.3
264 9606 9606.ENSP00000269305 9014 9014.A0A226P7S4 224.2
265 9606 9606.ENSP00000269305 283909 283909.R7UHV7 209.1
266 9606 9606.ENSP00000269305 6573 6573.A0A210QTK4 203.4
267 9606 9606.ENSP00000269305 7719 7719.ENSCINP00000003956 200.7
268 9606 9606.ENSP00000269305 37653 37653.A0A0L8G262 198.7
269 9606 9606.ENSP00000269305 225164 225164.V4A869 194.5
270 9606 9606.ENSP00000269305 6526 6526.A0A2C9K4K6 193.7
271 9606 9606.ENSP00000269305 7574 7574.A0A1S3IK29 192.2
272 9606 9606.ENSP00000269305 188477 188477.A0A433U4Z3 190.7
273 9606 9606.ENSP00000269305 51511 51511.ENSCSAVP00000016940 185.7
274 9606 9606.ENSP00000269305 10195 10195.A0A3M7Q6B9 184.5
275 9606 9606.ENSP00000269305 7668 7668.W4XZV7 175.3
276 9606 9606.ENSP00000269305 10228 10228.B3RZS6 162.2
277 9606 9606.ENSP00000269305 46731 46731.A0A3M6T6Q0 161.8
278 9606 9606.ENSP00000269305 287889 287889.A0A369SDE5 161.8
279 9606 9606.ENSP00000269305 407821 407821.A0A087U4M0 159.5
280 9606 9606.ENSP00000269305 50429 50429.A0A2B4RUK1 158.7
281 9606 9606.ENSP00000269305 6832 6832.A0A553NYY4 153.3
282 9606 9606.ENSP00000269305 307972 307972.A0A2G8L3N1 150.6
283 9606 9606.ENSP00000269305 418985 418985.A0A1V9Y190 149.8
284 9606 9606.ENSP00000269305 7070 7070.D7EJV6 144.1
285 9606 9606.ENSP00000269305 45351 45351.A7S162 142.9
286 9606 9606.ENSP00000269305 30538 30538.ENSVPAP00000011479 137.5
287 9606 9606.ENSP00000269305 6669 6669.E9FW11 135.2
288 9606 9606.ENSP00000269305 6945 6945.B7QF52 134.0
289 9606 9606.ENSP00000269305 6973 6973.A0A2P8XLZ2 134.0
290 9606 9606.ENSP00000269305 400727 400727.A0A2T7PT95 131.0
291 9606 9606.ENSP00000269305 35525 35525.A0A0P5UBN4 127.1
292 9606 9606.ENSP00000269305 126957 126957.T1INQ8 124.4
293 9606 9606.ENSP00000269305 7029 7029.J9JP39 123.2
294 9606 9606.ENSP00000269305 121224 121224.E0V916 122.5
295 9606 9606.ENSP00000269305 224129 224129.A0A1W4XBP0 121.7
296 9606 9606.ENSP00000269305 136037 136037.A0A067QKS2 120.9
297 9606 9606.ENSP00000269305 1661398 1661398.A0A482VJ27 120.9
298 9606 9606.ENSP00000269305 6412 6412.T1EZJ4 120.2
299 9606 9606.ENSP00000269305 121845 121845.A0A3Q0IPE1 113.6
300 9606 9606.ENSP00000269305 543379 543379.A0A232FJ48 112.8
301 9606 9606.ENSP00000269305 195883 195883.A0A482XMX2 110.2
302 9606 9606.ENSP00000269305 7425 7425.K7IV69 109.8
303 9606 9606.ENSP00000269305 597456 597456.A0A0L7RJB6 108.6
304 9606 9606.ENSP00000269305 67767 67767.A0A0J7L4T2 107.8
305 9606 9606.ENSP00000269305 81824 81824.A9UZX3 106.7
306 9606 9606.ENSP00000269305 568069 568069.A0A1J1HKW3 105.5
307 9606 9606.ENSP00000269305 166423 166423.A0A0N1IU42 105.1
308 9606 9606.ENSP00000269305 178035 178035.A0A154PH19 105.1
309 9606 9606.ENSP00000269305 94128 94128.A0A2A3EPF3 104.8
310 9606 9606.ENSP00000269305 7460 7460.A0A088AGC6 104.4
311 9606 9606.ENSP00000269305 268475 268475.A0A0V1H034 103.6
312 9606 9606.ENSP00000269305 6337 6337.A0A0V0XWZ0 103.6
313 9606 9606.ENSP00000269305 105785 105785.A0A2J7RT65 103.2
314 9606 9606.ENSP00000269305 268474 268474.A0A0V1MEE0 102.4
315 9606 9606.ENSP00000269305 13347 13347.A0A444TIG8 102.1
316 9606 9606.ENSP00000269305 13249 13249.T1HHW8 101.3
317 9606 9606.ENSP00000269305 70415 70415.A0A5S6Q6J5 100.5
318 9606 9606.ENSP00000269305 990121 990121.A0A0V0ZFM8 96.3
319 9606 9606.ENSP00000269305 45882 45882.A0A0V1D8F8 95.5
320 9606 9606.ENSP00000269305 92179 92179.A0A0V0WQ02 95.5
321 9606 9606.ENSP00000269305 144512 144512.A0A0V0U5U4 95.5
322 9606 9606.ENSP00000269305 667725 667725.A0A0L0G1F4 95.5
323 9606 9606.ENSP00000269305 6335 6335.A0A0V1LDC9 95.5
324 9606 9606.ENSP00000269305 610380 610380.E2B2V4 95.1
325 9606 9606.ENSP00000269305 104421 104421.E2APG2 94.7
326 9606 9606.ENSP00000269305 6689 6689.A0A3R7SPV9 94.7
327 9606 9606.ENSP00000269305 7209 7209.A0A1I7VBH3 94.7
328 9606 9606.ENSP00000269305 946362 946362.F2U8Q9 94.4
329 9606 9606.ENSP00000269305 181606 181606.A0A0V0V4L8 93.6
330 9606 9606.ENSP00000269305 92180 92180.A0A0V1P3X2 93.2
331 9606 9606.ENSP00000269305 77166 77166.N6UG92 92.8
332 9606 9606.ENSP00000269305 6336 6336.A0A0V0S9D5 92.0
333 9606 9606.ENSP00000269305 42155 42155.A0A0R3QQH5 90.9
334 9606 9606.ENSP00000269305 51028 51028.A0A0N4VCR4 90.5
335 9606 9606.ENSP00000269305 456900 456900.A0A195C1G0 89.7
336 9606 9606.ENSP00000269305 6334 6334.A0A0V1B7T2 89.7
337 9606 9606.ENSP00000269305 7176 7176.B0W6K7 89.0
338 9606 9606.ENSP00000269305 42156 42156.A0A3P7K7U3 88.2
339 9606 9606.ENSP00000269305 46835 46835.A0A504YVH4 87.4
340 9606 9606.ENSP00000269305 6183 6183.A0A3Q0KN56 87.4
341 9606 9606.ENSP00000269305 6280 6280.A0A0N4TIF7 87.4
342 9606 9606.ENSP00000269305 595528 595528.A0A0D2X0F0 86.3
343 9606 9606.ENSP00000269305 6282 6282.A0A044R7Z7 86.3
344 9606 9606.ENSP00000269305 6186 6186.A0A183JFI6 85.9
345 9606 9606.ENSP00000269305 60517 60517.A0A158R7U3 85.1
346 9606 9606.ENSP00000269305 1147741 1147741.A0A0R3RXL0 85.1
347 9606 9606.ENSP00000269305 6184 6184.A0A430QT47 84.7
348 9606 9606.ENSP00000269305 102285 102285.A0A0R3TMT8 84.3
349 9606 9606.ENSP00000269305 300112 300112.A0A4S2JFH1 84.3
350 9606 9606.ENSP00000269305 278856 278856.A0A212FJ15 84.0
351 9606 9606.ENSP00000269305 6182 6182.A0A4Z2CRH0 84.0
352 9606 9606.ENSP00000269305 6211 6211.A0A087VYM5 84.0
353 9606 9606.ENSP00000269305 6277 6277.A0A498SGZ8 84.0
354 9606 9606.ENSP00000269305 64791 64791.A0A151XH99 83.6
355 9606 9606.ENSP00000269305 6185 6185.A0A095C5C5 83.2
356 9606 9606.ENSP00000269305 6210 6210.W6U916 82.8
357 9606 9606.ENSP00000269305 7160 7160.A0A182GIF0 82.8
358 9606 9606.ENSP00000269305 7159 7159.Q171M1 81.6
359 9606 9606.ENSP00000269305 53468 53468.A0A0R3U6F0 80.9
360 9606 9606.ENSP00000269305 6293 6293.A0A1I8EB15 80.9
361 9606 9606.ENSP00000269305 6205 6205.A0A0R3WQW6 80.5
362 9606 9606.ENSP00000269305 79923 79923.A0A3R7GQP5 80.1
363 9606 9606.ENSP00000269305 147828 147828.A0A4S2LLK3 80.1
364 9606 9606.ENSP00000269305 6198 6198.A0A074ZCB0 80.1
365 9606 9606.ENSP00000269305 6216 6216.A0A0R3S7X9 79.7
366 9606 9606.ENSP00000269305 103372 103372.F4X756 79.0
367 9606 9606.ENSP00000269305 2015173 2015173.A0A026WHR3 79.0
368 9606 9606.ENSP00000269305 34720 34720.A0A195FHS0 78.6
369 9606 9606.ENSP00000269305 37001 37001.A0A1A9W1P0 78.6
370 9606 9606.ENSP00000269305 66420 66420.A0A0N1PE05 77.8
371 9606 9606.ENSP00000269305 168631 168631.A0A437B4Y2 77.8
372 9606 9606.ENSP00000269305 520822 520822.A0A195B173 77.8
373 9606 9606.ENSP00000269305 471704 471704.A0A195DK69 77.4
374 9606 9606.ENSP00000269305 12957 12957.A0A158ND95 77.4
375 9606 9606.ENSP00000269305 451379 451379.A0A0N5B0J2 77.0
376 9606 9606.ENSP00000269305 7398 7398.A0A1A9ZZR1 77.0
377 9606 9606.ENSP00000269305 29172 29172.A0A0D8Y6I2 75.9
378 9606 9606.ENSP00000269305 400682 400682.A0A1X7UD97 75.1
379 9606 9606.ENSP00000269305 201502 201502.A0A1A9YB05 74.7
380 9606 9606.ENSP00000269305 6265 6265.A0A0B2UVT7 74.3
381 9606 9606.ENSP00000269305 37546 37546.A0A1B0G0N7 73.6
382 9606 9606.ENSP00000269305 76193 76193.A0A0N0PCE7 72.0
383 9606 9606.ENSP00000269305 7395 7395.A0A1A9UZU7 72.0
384 9606 9606.ENSP00000269305 62324 62324.A0A182RCW2 70.5
385 9606 9606.ENSP00000269305 199890 199890.A0A182PA25 70.5
386 9606 9606.ENSP00000269305 158441 158441.A0A226DFT1 70.1
387 9606 9606.ENSP00000269305 69004 69004.A0A3F2YX69 69.3
388 9606 9606.ENSP00000269305 43151 43151.W5JCA5 68.9
389 9606 9606.ENSP00000269305 7167 7167.A0A182FR67 68.6
390 9606 9606.ENSP00000269305 334426 334426.A0A0R3Q2A0 67.0
391 9606 9606.ENSP00000269305 7102 7102.A0A2A4JNC2 67.0
392 9606 9606.ENSP00000269305 139723 139723.A0A182M5Y6 66.6
393 9606 9606.ENSP00000269305 6313 6313.A0A0K0DP08 66.6
394 9606 9606.ENSP00000269305 6290 6290.A0A0N4W3M7 66.2
395 9606 9606.ENSP00000269305 7168 7168.A0A182N4N0 65.9
396 9606 9606.ENSP00000269305 41427 41427.A0A182J735 65.9
397 9606 9606.ENSP00000269305 51031 51031.W2SWL6 65.5
398 9606 9606.ENSP00000269305 53326 53326.A0A016WYV3 65.5
399 9606 9606.ENSP00000269305 48709 48709.A0A1D2NAH9 65.1
400 9606 9606.ENSP00000269305 7165 7165.F5HK78 65.1
401 9606 9606.ENSP00000269305 7260 7260.B4N9F0 65.1
402 9606 9606.ENSP00000269305 1518534 1518534.A0A182LFH4 65.1
403 9606 9606.ENSP00000269305 74873 74873.A0A084W6W1 63.9
404 9606 9606.ENSP00000269305 112268 112268.A0A182VY33 63.9
405 9606 9606.ENSP00000269305 30066 30066.A0A182V190 63.9
406 9606 9606.ENSP00000269305 34691 34691.A0A182XID1 63.9
407 9606 9606.ENSP00000269305 42157 42157.A0A182ELX1 63.9
408 9606 9606.ENSP00000269305 46245 46245.A0A0R3NKT4 63.2
409 9606 9606.ENSP00000269305 29170 29170.A0A368F8M2 63.2
410 9606 9606.ENSP00000269305 318479 318479.A0A0N4UDB8 62.8
411 9606 9606.ENSP00000269305 7234 7234.B4GZG1 62.4
412 9606 9606.ENSP00000269305 30069 30069.A0A182Y7A0 61.6
413 9606 9606.ENSP00000269305 43041 43041.A0A182K2G4 60.5
414 9606 9606.ENSP00000269305 7217 7217.A0A0P8XYV2 59.3
415 9606 9606.ENSP00000269305 7227 7227.FBpp0083753 58.5
416 9606 9606.ENSP00000269305 7232 7232.A0A484BSX2 58.5
417 9606 9606.ENSP00000269305 387005 387005.A0A183H0Z1 57.0
418 9606 9606.ENSP00000269305 7222 7222.B4JSF4 57.0
419 9606 9606.ENSP00000269305 7238 7238.B4HFA1 57.0
420 9606 9606.ENSP00000269305 70667 70667.A0A183STT5 56.6
421 9606 9606.ENSP00000269305 7370 7370.A0A1I8N839 56.6
422 9606 9606.ENSP00000269305 35570 35570.A0A1I8QBH4 56.6
423 9606 9606.ENSP00000269305 7091 7091.E9JEI4 56.2
424 9606 9606.ENSP00000269305 27835 27835.A0A0N4YQF9 55.1
425 9606 9606.ENSP00000269305 7240 7240.B4R1C4 54.3
426 9606 9606.ENSP00000269305 34690 34690.A0A182UJE7 53.5
427 9606 9606.ENSP00000269305 7375 7375.A0A0L0CLI5 53.1
428 9606 9606.ENSP00000269305 7266 7266.A0A3B0JRN9 52.0
429 9606 9606.ENSP00000269305 30019 30019.A0A0M3QXK1 52.0
430 9606 9606.ENSP00000269305 947166 947166.A0A1D1VBG3 51.6
431 9606 9606.ENSP00000269305 7244 7244.B4MB49 51.2
432 9606 9606.ENSP00000269305 54126 54126.H3FK89 50.8If you want to find the best hits (based on bit score) of a specific protein in other species, you’d use the get_homologs_besthits() method.
# Get best hits of tp53 (human) in other species
string_db$get_homologs_besthits(tp53) |> head() X9606 X9606.ENSP00000269305 X9597 X9597.ENSPPAP00000011040 X815.8
1 9606 9606.ENSP00000269305 9598 9598.ENSPTRP00000014836 815.8
2 9606 9606.ENSP00000269305 9606 9606.ENSP00000269305 815.8
3 9606 9606.ENSP00000269305 9601 9601.ENSPPYP00000008923 799.3
4 9606 9606.ENSP00000269305 61621 61621.ENSRBIP00000028632 786.6
5 9606 9606.ENSP00000269305 61622 61622.ENSRROP00000026843 786.6
6 9606 9606.ENSP00000269305 591936 591936.ENSPTEP00000010553 785.8You can also extract the best hit in a specific species by specifying the target species Taxonomy ID in argument target_species_id.
# Get best hit of tp53 in the mouse genome (Mus musculus, TaxID = 10090)
string_db$get_homologs_besthits(tp53, target_species_id = 10090)[1] X9606 X9606.ENSP00000269305
[3] X10090 X10090.ENSMUSP00000104298
[5] X598.2
<0 rows> (or 0-length row.names)Find the best hits of ATM in other species, then answer the questions below:
- Which species has the most similar protein to ATM?
- Which species has the least similar protein to ATM?
- How do you interpret these results?
Session information
This chapter was created under the following conditions:
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os Ubuntu 22.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Europe/Brussels
date 2024-05-27
pandoc 3.1.1 @ /usr/lib/rstudio/resources/app/bin/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
BiocManager 1.30.22 2023-08-08 [1] CRAN (R 4.3.2)
BiocStyle 2.30.0 2023-10-24 [1] Bioconductor
bit 4.0.5 2022-11-15 [1] CRAN (R 4.3.2)
bit64 4.0.5 2020-08-30 [1] CRAN (R 4.3.2)
bitops 1.0-7 2021-04-24 [1] CRAN (R 4.3.2)
blob 1.2.4 2023-03-17 [1] CRAN (R 4.3.2)
cachem 1.0.8 2023-05-01 [1] CRAN (R 4.3.2)
caTools 1.18.2 2021-03-28 [1] CRAN (R 4.3.2)
chron 2.3-61 2023-05-02 [1] CRAN (R 4.3.2)
cli 3.6.2 2023-12-11 [1] CRAN (R 4.3.2)
colorspace 2.1-0 2023-01-23 [1] CRAN (R 4.3.2)
curl 5.2.0 2023-12-08 [1] CRAN (R 4.3.2)
DBI 1.2.1 2024-01-12 [1] CRAN (R 4.3.2)
digest 0.6.34 2024-01-11 [1] CRAN (R 4.3.2)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.3.2)
evaluate 0.23 2023-11-01 [1] CRAN (R 4.3.2)
fansi 1.0.6 2023-12-08 [1] CRAN (R 4.3.2)
fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.3.2)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.3.2)
generics 0.1.3 2022-07-05 [1] CRAN (R 4.3.2)
ggplot2 * 3.5.0 2024-02-23 [1] CRAN (R 4.3.2)
glue 1.7.0 2024-01-09 [1] CRAN (R 4.3.2)
gplots 3.1.3.1 2024-02-02 [1] CRAN (R 4.3.2)
gsubfn 0.7 2018-03-16 [1] CRAN (R 4.3.2)
gtable 0.3.4 2023-08-21 [1] CRAN (R 4.3.2)
gtools 3.9.5 2023-11-20 [1] CRAN (R 4.3.2)
hash 2.2.6.3 2023-08-19 [1] CRAN (R 4.3.2)
hms 1.1.3 2023-03-21 [1] CRAN (R 4.3.2)
htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.3.2)
htmlwidgets 1.6.4 2023-12-06 [1] CRAN (R 4.3.2)
httr 1.4.7 2023-08-15 [1] CRAN (R 4.3.2)
igraph * 2.0.1.1 2024-01-30 [1] CRAN (R 4.3.2)
jsonlite 1.8.8 2023-12-04 [1] CRAN (R 4.3.2)
KernSmooth 2.23-22 2023-07-10 [4] CRAN (R 4.3.1)
knitr 1.45 2023-10-30 [1] CRAN (R 4.3.2)
lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.3.2)
lubridate * 1.9.3 2023-09-27 [1] CRAN (R 4.3.2)
magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.3.2)
memoise 2.0.1 2021-11-26 [1] CRAN (R 4.3.2)
munsell 0.5.0 2018-06-12 [1] CRAN (R 4.3.2)
pillar 1.9.0 2023-03-22 [1] CRAN (R 4.3.2)
pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.3.2)
plotrix 3.8-4 2023-11-10 [1] CRAN (R 4.3.2)
plyr 1.8.9 2023-10-02 [1] CRAN (R 4.3.2)
png 0.1-8 2022-11-29 [1] CRAN (R 4.3.2)
proto 1.0.0 2016-10-29 [1] CRAN (R 4.3.2)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.3.2)
R6 2.5.1 2021-08-19 [1] CRAN (R 4.3.2)
RColorBrewer 1.1-3 2022-04-03 [1] CRAN (R 4.3.2)
Rcpp 1.0.12 2024-01-09 [1] CRAN (R 4.3.2)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.3.2)
rlang 1.1.3 2024-01-10 [1] CRAN (R 4.3.2)
rmarkdown 2.25 2023-09-18 [1] CRAN (R 4.3.2)
RSQLite 2.3.5 2024-01-21 [1] CRAN (R 4.3.2)
rstudioapi 0.15.0 2023-07-07 [1] CRAN (R 4.3.2)
scales 1.3.0 2023-11-28 [1] CRAN (R 4.3.2)
sessioninfo 1.2.2 2021-12-06 [1] CRAN (R 4.3.2)
sqldf 0.4-11 2017-06-28 [1] CRAN (R 4.3.2)
STRINGdb * 2.14.3 2024-03-13 [1] Bioconductor 3.18 (R 4.3.2)
stringi 1.8.3 2023-12-11 [1] CRAN (R 4.3.2)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.3.2)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.3.2)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.3.2)
tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.3.2)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.3.2)
timechange 0.3.0 2024-01-18 [1] CRAN (R 4.3.2)
tzdb 0.4.0 2023-05-12 [1] CRAN (R 4.3.2)
utf8 1.2.4 2023-10-22 [1] CRAN (R 4.3.2)
vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.3.2)
withr 3.0.0 2024-01-16 [1] CRAN (R 4.3.2)
xfun 0.42 2024-02-08 [1] CRAN (R 4.3.2)
yaml 2.3.8 2023-12-11 [1] CRAN (R 4.3.2)
[1] /home/faalm/R/x86_64-pc-linux-gnu-library/4.3
[2] /usr/local/lib/R/site-library
[3] /usr/lib/R/site-library
[4] /usr/lib/R/library
──────────────────────────────────────────────────────────────────────────────