library(tidyverse)
library(here)2 Data processing with the tidyverse
The tidyverse package is an “umbrella-package” that installs several useful packages for data analysis which work well together, such as tidyr, dplyr, ggplot2, tibble, etc. These packages help us to work and interact with the data. They allow us to do many things with your data, such as subsetting, transforming, visualizing, etc.
Let’s start by loading the tidyverse package:
2.1 Goals of this lesson
At the end of this lesson, you will be able to:
- read tabular data from .csv/.tsv/.txt/ files
- filter your data set to select particular rows and columns
- add columns
- summarize data
- combine different data frames
- reshape data from long to wide, and back
2.2 The readr package: reading files
The data we will use in this lesson are stored in data/rnaseq.csv. This is a comma-separated file, which means it contains a table in which columns are separated by commas. To read .csv files, we will use the function read_csv() from the readr package.
# Read csv file
rna <- read_csv(here("data", "rnaseq.csv"))
# Inspect the data
head(rna)# A tibble: 6 × 19
gene sample expression organism age sex infection strain time tissue
<chr> <chr> <dbl> <chr> <dbl> <chr> <chr> <chr> <dbl> <chr>
1 Asl GSM2545… 1170 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
2 Apod GSM2545… 36194 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
3 Cyp2d22 GSM2545… 4060 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
4 Klk6 GSM2545… 287 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
5 Fcrls GSM2545… 85 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
6 Slc2a4 GSM2545… 782 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
# ℹ 9 more variables: mouse <dbl>, ENTREZID <dbl>, product <chr>,
# ensembl_gene_id <chr>, external_synonym <chr>, chromosome_name <chr>,
# gene_biotype <chr>, phenotype_description <chr>,
# hsapiens_homolog_associated_gene_name <chr>Notice that the class of the data is now referred to as a tibble.
Tibbles tweak some of the behaviors of the data frame objects we introduced in the previously. The data structure is very similar to a data frame. For our purposes the only differences are that:
It displays the data type of each column under its name. Note that <
dbl> is a data type defined to hold numeric values with decimal points.It only prints the first few rows of data and only as many columns as fit on one screen.
2.3 The dplyr package: filtering, extending, and summarizing data
The dplyr is one of the most main packages of the tidyverse, and it can be used to perform all sorts of day-to-day data processing. Here, we are going to learn some of the most common dplyr functions:
select(): subset columnsfilter(): subset rows on conditionsmutate(): create new columns by using information from other columnsgroup_by()andsummarise(): create summary statistics on grouped dataarrange(): sort resultscount(): count discrete values
2.3.1 Selecting columns and filtering rows
To select columns of a data frame, use select(). The first argument to this function is the data frame (rna), and the subsequent arguments are the columns to keep.
select(rna, gene, sample, tissue, expression)# A tibble: 32,428 × 4
gene sample tissue expression
<chr> <chr> <chr> <dbl>
1 Asl GSM2545336 Cerebellum 1170
2 Apod GSM2545336 Cerebellum 36194
3 Cyp2d22 GSM2545336 Cerebellum 4060
4 Klk6 GSM2545336 Cerebellum 287
5 Fcrls GSM2545336 Cerebellum 85
6 Slc2a4 GSM2545336 Cerebellum 782
7 Exd2 GSM2545336 Cerebellum 1619
8 Gjc2 GSM2545336 Cerebellum 288
9 Plp1 GSM2545336 Cerebellum 43217
10 Gnb4 GSM2545336 Cerebellum 1071
# ℹ 32,418 more rowsTo select all columns except certain ones, put a “-” in front of the variable to exclude it. For example, to select all columns but tissue and organism, you’d use:
select(rna, -tissue, -organism)# A tibble: 32,428 × 17
gene sample expression age sex infection strain time mouse ENTREZID
<chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 Asl GSM2545… 1170 8 Fema… Influenz… C57BL… 8 14 109900
2 Apod GSM2545… 36194 8 Fema… Influenz… C57BL… 8 14 11815
3 Cyp2d22 GSM2545… 4060 8 Fema… Influenz… C57BL… 8 14 56448
4 Klk6 GSM2545… 287 8 Fema… Influenz… C57BL… 8 14 19144
5 Fcrls GSM2545… 85 8 Fema… Influenz… C57BL… 8 14 80891
6 Slc2a4 GSM2545… 782 8 Fema… Influenz… C57BL… 8 14 20528
7 Exd2 GSM2545… 1619 8 Fema… Influenz… C57BL… 8 14 97827
8 Gjc2 GSM2545… 288 8 Fema… Influenz… C57BL… 8 14 118454
9 Plp1 GSM2545… 43217 8 Fema… Influenz… C57BL… 8 14 18823
10 Gnb4 GSM2545… 1071 8 Fema… Influenz… C57BL… 8 14 14696
# ℹ 32,418 more rows
# ℹ 7 more variables: product <chr>, ensembl_gene_id <chr>,
# external_synonym <chr>, chromosome_name <chr>, gene_biotype <chr>,
# phenotype_description <chr>, hsapiens_homolog_associated_gene_name <chr>To choose rows based on specific criteria, use filter():
filter(rna, sex == "Male")# A tibble: 14,740 × 19
gene sample expression organism age sex infection strain time tissue
<chr> <chr> <dbl> <chr> <dbl> <chr> <chr> <chr> <dbl> <chr>
1 Asl GSM254… 626 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
2 Apod GSM254… 13021 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
3 Cyp2d22 GSM254… 2171 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
4 Klk6 GSM254… 448 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
5 Fcrls GSM254… 180 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
6 Slc2a4 GSM254… 313 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
7 Exd2 GSM254… 2366 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
8 Gjc2 GSM254… 310 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
9 Plp1 GSM254… 53126 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
10 Gnb4 GSM254… 1355 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
# ℹ 14,730 more rows
# ℹ 9 more variables: mouse <dbl>, ENTREZID <dbl>, product <chr>,
# ensembl_gene_id <chr>, external_synonym <chr>, chromosome_name <chr>,
# gene_biotype <chr>, phenotype_description <chr>,
# hsapiens_homolog_associated_gene_name <chr>filter(rna, sex == "Male" & infection == "NonInfected")# A tibble: 4,422 × 19
gene sample expression organism age sex infection strain time tissue
<chr> <chr> <dbl> <chr> <dbl> <chr> <chr> <chr> <dbl> <chr>
1 Asl GSM254… 535 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
2 Apod GSM254… 13668 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
3 Cyp2d22 GSM254… 2008 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
4 Klk6 GSM254… 1101 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
5 Fcrls GSM254… 375 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
6 Slc2a4 GSM254… 249 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
7 Exd2 GSM254… 3126 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
8 Gjc2 GSM254… 791 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
9 Plp1 GSM254… 98658 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
10 Gnb4 GSM254… 2437 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
# ℹ 4,412 more rows
# ℹ 9 more variables: mouse <dbl>, ENTREZID <dbl>, product <chr>,
# ensembl_gene_id <chr>, external_synonym <chr>, chromosome_name <chr>,
# gene_biotype <chr>, phenotype_description <chr>,
# hsapiens_homolog_associated_gene_name <chr>Now, let’s imagine we are interested in the human homologs of the mouse genes analyzed in this dataset. This information can be found in the last column of the rna tibble, named hsapiens_homolog_associated_gene_name. To visualise it easily, we will create a new table containing just the 2 columns gene and hsapiens_homolog_associated_gene_name.
genes <- select(rna, gene, hsapiens_homolog_associated_gene_name)
genes# A tibble: 32,428 × 2
gene hsapiens_homolog_associated_gene_name
<chr> <chr>
1 Asl ASL
2 Apod APOD
3 Cyp2d22 CYP2D6
4 Klk6 KLK6
5 Fcrls FCRL2
6 Slc2a4 SLC2A4
7 Exd2 EXD2
8 Gjc2 GJC2
9 Plp1 PLP1
10 Gnb4 GNB4
# ℹ 32,418 more rowsA very nice thing about tidyverse verbs (functions) is that they can be executed one after the other by using the pipe operator (|> or %>%). In practice, that means you don’t have to create intermediate objects for complicated subsetting operations that involve multiple steps. The pipe operator is often read as and then in the sense that you apply a function A and then you pass the output to function B, and then you pass the output to function C, and so on and so forth. For example:
- 1
-
Take the object
rna, and then - 2
-
filter it to keep only rows that have “Male” in the column
sex, and then - 3
-
select the columns
gene,sample,tissue,expression
# A tibble: 14,740 × 4
gene sample tissue expression
<chr> <chr> <chr> <dbl>
1 Asl GSM2545340 Cerebellum 626
2 Apod GSM2545340 Cerebellum 13021
3 Cyp2d22 GSM2545340 Cerebellum 2171
4 Klk6 GSM2545340 Cerebellum 448
5 Fcrls GSM2545340 Cerebellum 180
6 Slc2a4 GSM2545340 Cerebellum 313
7 Exd2 GSM2545340 Cerebellum 2366
8 Gjc2 GSM2545340 Cerebellum 310
9 Plp1 GSM2545340 Cerebellum 53126
10 Gnb4 GSM2545340 Cerebellum 1355
# ℹ 14,730 more rows- Using pipes, subset the
rnatibble to keep observations that match the following criteria:
Female mice
Time point 0
Expression higher than 50000
Then, select the columns
gene,sample,time,expression, andage.
Filter
rnato keep observations for infected samples, then count the number of rows. Hint: use theunique()function to see all the unique observations in a column, and thenrow()function the get the number of rows in a data frame/tibble.Do the same as above, but now keep only non-infected samples.
2.3.2 Adding new columns
Frequently you’ll want to create new columns based on the values of existing columns, for example to do unit conversions, or to find the ratio of values in two columns. For this, we’ll use mutate().
For example, the column time contains the time in days. Let’s create a new column named time_hours that contains the time in hours.
rna |>
mutate(time_hours = time * 24) |>
select(time, time_hours)# A tibble: 32,428 × 2
time time_hours
<dbl> <dbl>
1 8 192
2 8 192
3 8 192
4 8 192
5 8 192
6 8 192
7 8 192
8 8 192
9 8 192
10 8 192
# ℹ 32,418 more rowsYou can also create a second new column based on the first new column within the same call of mutate():
rna |>
mutate(
time_hours = time * 24,
time_minutes = time_hours * 60
) |>
select(time, time_hours, time_minutes)# A tibble: 32,428 × 3
time time_hours time_minutes
<dbl> <dbl> <dbl>
1 8 192 11520
2 8 192 11520
3 8 192 11520
4 8 192 11520
5 8 192 11520
6 8 192 11520
7 8 192 11520
8 8 192 11520
9 8 192 11520
10 8 192 11520
# ℹ 32,418 more rowsCreate a new data frame from the rna data that meets the following criteria: contains only the gene, chromosome_name, phenotype_description, sample, and expression columns. The expression values should be log-transformed. This data frame must only contain genes located on sex chromosomes, associated with a phenotype_description, and with a log expression higher than 5.
Hint: think about how the commands should be ordered to produce this data frame!
2.3.3 Summarizing grouped data
Many data analysis tasks can be approached using the split-apply-combine paradigm: split the data into groups, apply some analysis to each group, and then combine the results. dplyr makes this very easy through the use of the group_by() function.
rna %>%
group_by(gene)# A tibble: 32,428 × 19
# Groups: gene [1,474]
gene sample expression organism age sex infection strain time tissue
<chr> <chr> <dbl> <chr> <dbl> <chr> <chr> <chr> <dbl> <chr>
1 Asl GSM254… 1170 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
2 Apod GSM254… 36194 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
3 Cyp2d22 GSM254… 4060 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
4 Klk6 GSM254… 287 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
5 Fcrls GSM254… 85 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
6 Slc2a4 GSM254… 782 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
7 Exd2 GSM254… 1619 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
8 Gjc2 GSM254… 288 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
9 Plp1 GSM254… 43217 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
10 Gnb4 GSM254… 1071 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
# ℹ 32,418 more rows
# ℹ 9 more variables: mouse <dbl>, ENTREZID <dbl>, product <chr>,
# ensembl_gene_id <chr>, external_synonym <chr>, chromosome_name <chr>,
# gene_biotype <chr>, phenotype_description <chr>,
# hsapiens_homolog_associated_gene_name <chr>The group_by() function doesn’t perform any data processing, it groups the data into subsets: in the example above, our initial tibble of 32428 observations is split into 1474 groups based on the gene variable.
Once the data has been grouped, subsequent operations will be applied on each group independently by using the summarise() function. For example, to compute the mean expression by gene, you’d do:
rna |>
group_by(gene) |>
summarise(mean_expression = mean(expression))# A tibble: 1,474 × 2
gene mean_expression
<chr> <dbl>
1 AI504432 1053.
2 AW046200 131.
3 AW551984 295.
4 Aamp 4751.
5 Abca12 4.55
6 Abcc8 2498.
7 Abhd14a 525.
8 Abi2 4909.
9 Abi3bp 1002.
10 Abl2 2124.
# ℹ 1,464 more rowsWe could also calculate the mean expression levels of all genes in each sample:
rna |>
group_by(sample) |>
summarise(mean_expression = mean(expression))# A tibble: 22 × 2
sample mean_expression
<chr> <dbl>
1 GSM2545336 2062.
2 GSM2545337 1766.
3 GSM2545338 1668.
4 GSM2545339 1696.
5 GSM2545340 1682.
6 GSM2545341 1638.
7 GSM2545342 1594.
8 GSM2545343 2107.
9 GSM2545344 1712.
10 GSM2545345 1700.
# ℹ 12 more rowsBut we can can also group by multiple columns:
rna |>
group_by(gene, infection, time) |>
summarise(mean_expression = mean(expression))# A tibble: 4,422 × 4
# Groups: gene, infection [2,948]
gene infection time mean_expression
<chr> <chr> <dbl> <dbl>
1 AI504432 InfluenzaA 4 1104.
2 AI504432 InfluenzaA 8 1014
3 AI504432 NonInfected 0 1034.
4 AW046200 InfluenzaA 4 152.
5 AW046200 InfluenzaA 8 81
6 AW046200 NonInfected 0 155.
7 AW551984 InfluenzaA 4 302.
8 AW551984 InfluenzaA 8 342.
9 AW551984 NonInfected 0 238
10 Aamp InfluenzaA 4 4870
# ℹ 4,412 more rowsCalculate the mean and median expression of all genes.
Calculate the mean expression level of gene “Dok3” by timepoints.
2.3.4 Counting observations per group
When working with data, we often want to know the number of observations found for each factor or combination of factors. For this task, dplyr provides count(). For example, if we wanted to count the number of rows of data for each infected and non-infected samples, we would do:
rna |>
count(infection)# A tibble: 2 × 2
infection n
<chr> <int>
1 InfluenzaA 22110
2 NonInfected 10318If we wanted to count a combination of factors, such as infection and time, we would specify the first and the second factor as the arguments of count():
rna |>
count(infection, time)# A tibble: 3 × 3
infection time n
<chr> <dbl> <int>
1 InfluenzaA 4 11792
2 InfluenzaA 8 10318
3 NonInfected 0 10318It is sometimes useful to sort the result to facilitate the comparisons. We can use arrange() to sort the table. For instance, we might want to arrange the table above by time:
# Ascending order
rna |>
count(infection, time) |>
arrange(time)# A tibble: 3 × 3
infection time n
<chr> <dbl> <int>
1 NonInfected 0 10318
2 InfluenzaA 4 11792
3 InfluenzaA 8 10318# Descending order
rna |>
count(infection, time) |>
arrange(-time)# A tibble: 3 × 3
infection time n
<chr> <dbl> <int>
1 InfluenzaA 8 10318
2 InfluenzaA 4 11792
3 NonInfected 0 10318How many genes were analyzed in each sample?
Use
group_by()andsummarise()to evaluate the sequencing depth (the sum of all counts) in each sample. Which sample has the highest sequencing depth?Pick one sample and evaluate the number of genes by biotype.
Identify genes associated with the “abnormal DNA methylation” phenotype description, and calculate their mean expression (in log) at time 0, time 4 and time 8.
2.3.5 Joining tables
In many real-life situations, data are spread across multiple tables. A common example in transcriptomics is to have gene expression in one table, and functional annotation for each gene in another table. In these cases, one may want to combine the two tables based on a column in common.
Joining tables can be done with the dplyr functions inner_join(), full_join(), left_join(), and right_join(), each of which are exemplified in the figure below:
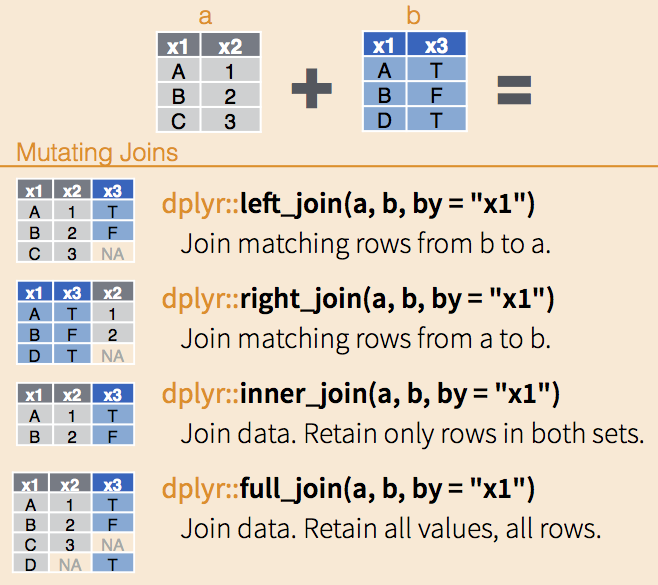
dplyr joinsTo demonstrate that, let’s first create a data frame with the mean expression of the genes Asl, Apod, and Klk6.
# Mean expression of the genes `Asl`, `Apod`, and Klk6
mean_exp <- rna |>
filter(gene %in% c("Asl", "Apod", "Klk6")) |>
group_by(gene) |>
summarise(mean_exp = mean(expression))Next, we will load a table with genes and their descriptions, available in data/gene_descriptions.csv.
# Read data
descriptions <- read_csv(here("data", "gene_descriptions.csv"))
# Inspect data
head(descriptions)# A tibble: 6 × 2
gene gene_description
<chr> <chr>
1 Cyp2d22 cytochrome P450, family 2, subfamily d, polypeptide 22 [Source:MGI Sy…
2 Klk6 kallikrein related-peptidase 6 [Source:MGI Symbol;Acc:MGI:1343166]
3 Fcrls Fc receptor-like S, scavenger receptor [Source:MGI Symbol;Acc:MGI:193…
4 Plp1 proteolipid protein (myelin) 1 [Source:MGI Symbol;Acc:MGI:97623]
5 Exd2 exonuclease 3'-5' domain containing 2 [Source:MGI Symbol;Acc:MGI:1922…
6 Apod apolipoprotein D [Source:MGI Symbol;Acc:MGI:88056] Now, we will combine the two tables so that we have the mean expression for each gene + gene descriptions in a single table. To combine the two tables while keeping only genes present in the column gene of both tables, we will use the function inner_join().
inner_join(mean_exp, descriptions)# A tibble: 3 × 3
gene mean_exp gene_description
<chr> <dbl> <chr>
1 Apod 18968. apolipoprotein D [Source:MGI Symbol;Acc:MGI:88056]
2 Asl 708. argininosuccinate lyase [Source:MGI Symbol;Acc:MGI:88084]
3 Klk6 554. kallikrein related-peptidase 6 [Source:MGI Symbol;Acc:MGI:1343…Join the tables mean_exp and descriptions using the function full_join(). How does the output change?
2.4 The tidyr package: from long to wide, and vice versa
In the rna tibble, the rows contain expression values (the unit) that are associated with a combination of 2 other variables: gene and sample.
All the other columns correspond to variables describing either the sample (organism, age, sex, …) or the gene (gene_biotype, ENTREZ_ID, product, …). The variables that don’t change with genes or with samples will have the same value in all the rows.
rna |>
arrange(gene)# A tibble: 32,428 × 19
gene sample expression organism age sex infection strain time tissue
<chr> <chr> <dbl> <chr> <dbl> <chr> <chr> <chr> <dbl> <chr>
1 AI504432 GSM25… 1230 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
2 AI504432 GSM25… 1085 Mus mus… 8 Fema… NonInfec… C57BL… 0 Cereb…
3 AI504432 GSM25… 969 Mus mus… 8 Fema… NonInfec… C57BL… 0 Cereb…
4 AI504432 GSM25… 1284 Mus mus… 8 Fema… Influenz… C57BL… 4 Cereb…
5 AI504432 GSM25… 966 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
6 AI504432 GSM25… 918 Mus mus… 8 Male Influenz… C57BL… 8 Cereb…
7 AI504432 GSM25… 985 Mus mus… 8 Fema… Influenz… C57BL… 8 Cereb…
8 AI504432 GSM25… 972 Mus mus… 8 Male NonInfec… C57BL… 0 Cereb…
9 AI504432 GSM25… 1000 Mus mus… 8 Fema… Influenz… C57BL… 4 Cereb…
10 AI504432 GSM25… 816 Mus mus… 8 Male Influenz… C57BL… 4 Cereb…
# ℹ 32,418 more rows
# ℹ 9 more variables: mouse <dbl>, ENTREZID <dbl>, product <chr>,
# ensembl_gene_id <chr>, external_synonym <chr>, chromosome_name <chr>,
# gene_biotype <chr>, phenotype_description <chr>,
# hsapiens_homolog_associated_gene_name <chr>This structure is called a long-format, as one column contains all the values, and other column(s) list(s) the context of the value.
In certain cases, the long-format is not really “human-readable”, and another format, a wide-format is preferred, as a more compact way of representing the data. This is typically the case with gene expression values that scientists are used to look as matrices, were rows represent genes and columns represent samples.
In this format, it would therefore become straightforward to explore the relationship between the gene expression levels within, and between, the samples.
# A tibble: 1,474 × 23
gene GSM2545336 GSM2545337 GSM2545338 GSM2545339 GSM2545340 GSM2545341
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Asl 1170 361 400 586 626 988
2 Apod 36194 10347 9173 10620 13021 29594
3 Cyp2d22 4060 1616 1603 1901 2171 3349
4 Klk6 287 629 641 578 448 195
5 Fcrls 85 233 244 237 180 38
6 Slc2a4 782 231 248 265 313 786
7 Exd2 1619 2288 2235 2513 2366 1359
8 Gjc2 288 595 568 551 310 146
9 Plp1 43217 101241 96534 58354 53126 27173
10 Gnb4 1071 1791 1867 1430 1355 798
# ℹ 1,464 more rows
# ℹ 16 more variables: GSM2545342 <dbl>, GSM2545343 <dbl>, GSM2545344 <dbl>,
# GSM2545345 <dbl>, GSM2545346 <dbl>, GSM2545347 <dbl>, GSM2545348 <dbl>,
# GSM2545349 <dbl>, GSM2545350 <dbl>, GSM2545351 <dbl>, GSM2545352 <dbl>,
# GSM2545353 <dbl>, GSM2545354 <dbl>, GSM2545362 <dbl>, GSM2545363 <dbl>,
# GSM2545380 <dbl>To convert the gene expression values from rna into a wide format, we need to create a new table where the values of the sample column would become the names of column variables.
The key point here is that we are still following a tidy data structure, but we have reshaped the data according to the observations of interest: expression levels per gene instead of recording them per gene and per sample.
Reshaping data from long to wide format can be performed with the function pivot_wider() from tidyr. For example:
rna_wide <- rna |>
1 select(gene, sample, expression) |>
pivot_wider(
2 names_from = sample,
3 values_from = expression
)
rna_wide- 1
-
Select columns
gene,sample, andexpression - 2
-
Columns names of the wide table will be the values of the
samplecolumn - 3
-
Values in the wide table will be the values of the
expressioncolumn
# A tibble: 1,474 × 23
gene GSM2545336 GSM2545337 GSM2545338 GSM2545339 GSM2545340 GSM2545341
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Asl 1170 361 400 586 626 988
2 Apod 36194 10347 9173 10620 13021 29594
3 Cyp2d22 4060 1616 1603 1901 2171 3349
4 Klk6 287 629 641 578 448 195
5 Fcrls 85 233 244 237 180 38
6 Slc2a4 782 231 248 265 313 786
7 Exd2 1619 2288 2235 2513 2366 1359
8 Gjc2 288 595 568 551 310 146
9 Plp1 43217 101241 96534 58354 53126 27173
10 Gnb4 1071 1791 1867 1430 1355 798
# ℹ 1,464 more rows
# ℹ 16 more variables: GSM2545342 <dbl>, GSM2545343 <dbl>, GSM2545344 <dbl>,
# GSM2545345 <dbl>, GSM2545346 <dbl>, GSM2545347 <dbl>, GSM2545348 <dbl>,
# GSM2545349 <dbl>, GSM2545350 <dbl>, GSM2545351 <dbl>, GSM2545352 <dbl>,
# GSM2545353 <dbl>, GSM2545354 <dbl>, GSM2545362 <dbl>, GSM2545363 <dbl>,
# GSM2545380 <dbl>To reshape data back to the long format, we would use the function pivot_longer().
rna_long <- rna_wide |>
pivot_longer(
1 -gene,
2 names_to = "sample",
3 values_to = "expression"
)
rna_long- 1
-
Use all columns except
gene. - 2
- Column names of the wide table will become a variable named sample
- 3
- Values in the wide table will become a variable named expression
# A tibble: 32,428 × 3
gene sample expression
<chr> <chr> <dbl>
1 Asl GSM2545336 1170
2 Asl GSM2545337 361
3 Asl GSM2545338 400
4 Asl GSM2545339 586
5 Asl GSM2545340 626
6 Asl GSM2545341 988
7 Asl GSM2545342 836
8 Asl GSM2545343 535
9 Asl GSM2545344 586
10 Asl GSM2545345 597
# ℹ 32,418 more rowsStarting from the rna table, use the pivot_wider() function to create a wide-format table giving the gene expression levels in each mouse. Then use the pivot_longer() function to restore a long-format table.
2.5 Exporting data
Finally, to export tabular data, we will use the write_* functions from the tidyr package. Here, we will export the tibble rna_wide to a .csv file. For that, we will use the function write_csv().
write_csv(rna_wide, file = here("output", "rna_wide.csv"))