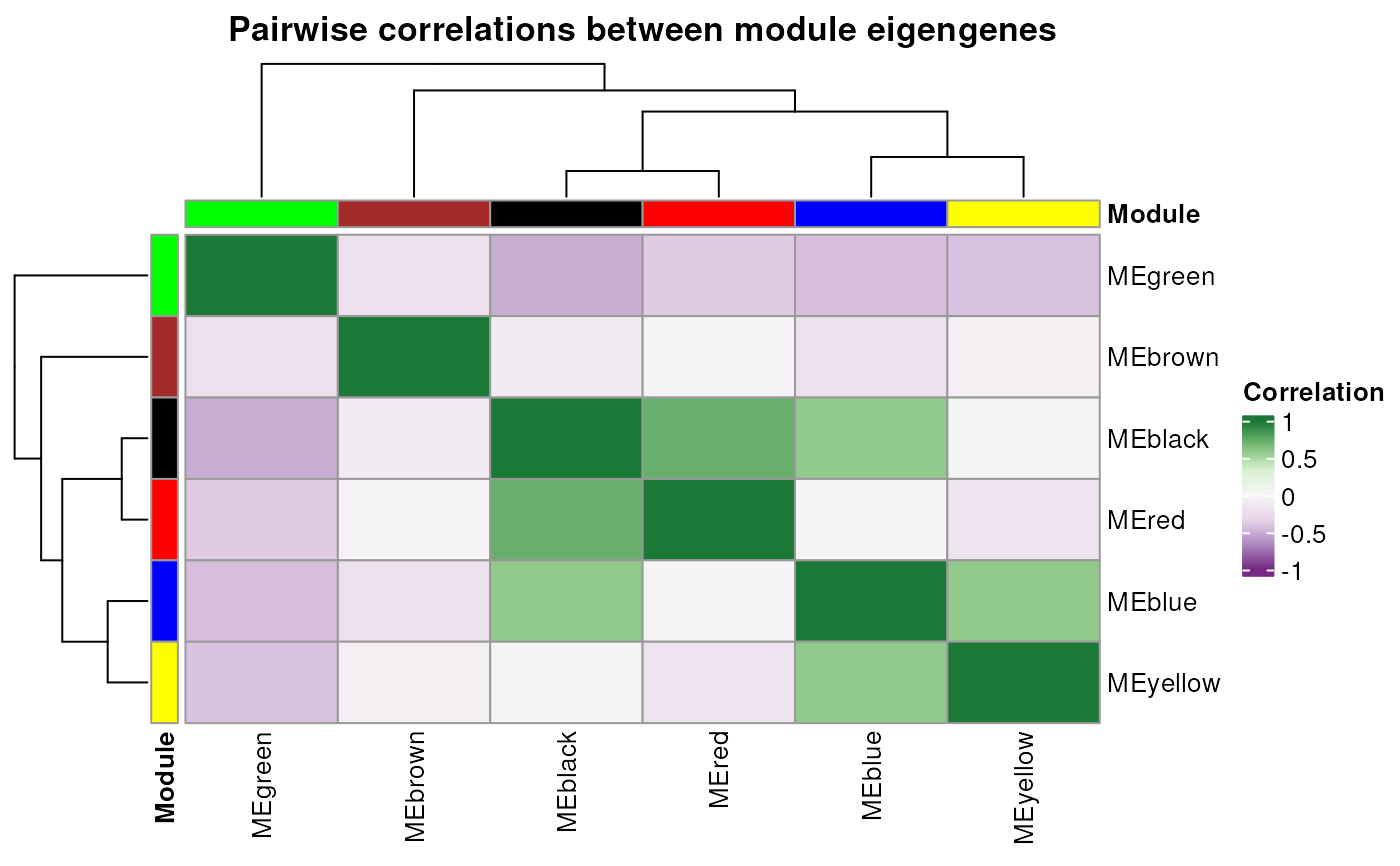
Usage
plot_eigengene_network(gcn, palette = "PRGn")
Arguments
- gcn
List object returned by exp2gcn.
- palette
Character indicating the name of the RColorBrewer palette
to use. Default: "PRGn".
Value
A base plot with the eigengene network
Examples
data(filt.se)
gcn <- exp2gcn(filt.se, SFTpower = 18, cor_method = "pearson")
#> ..connectivity..
#> ..matrix multiplication (system BLAS)..
#> ..normalization..
#> ..done.
plot_eigengene_network(gcn)