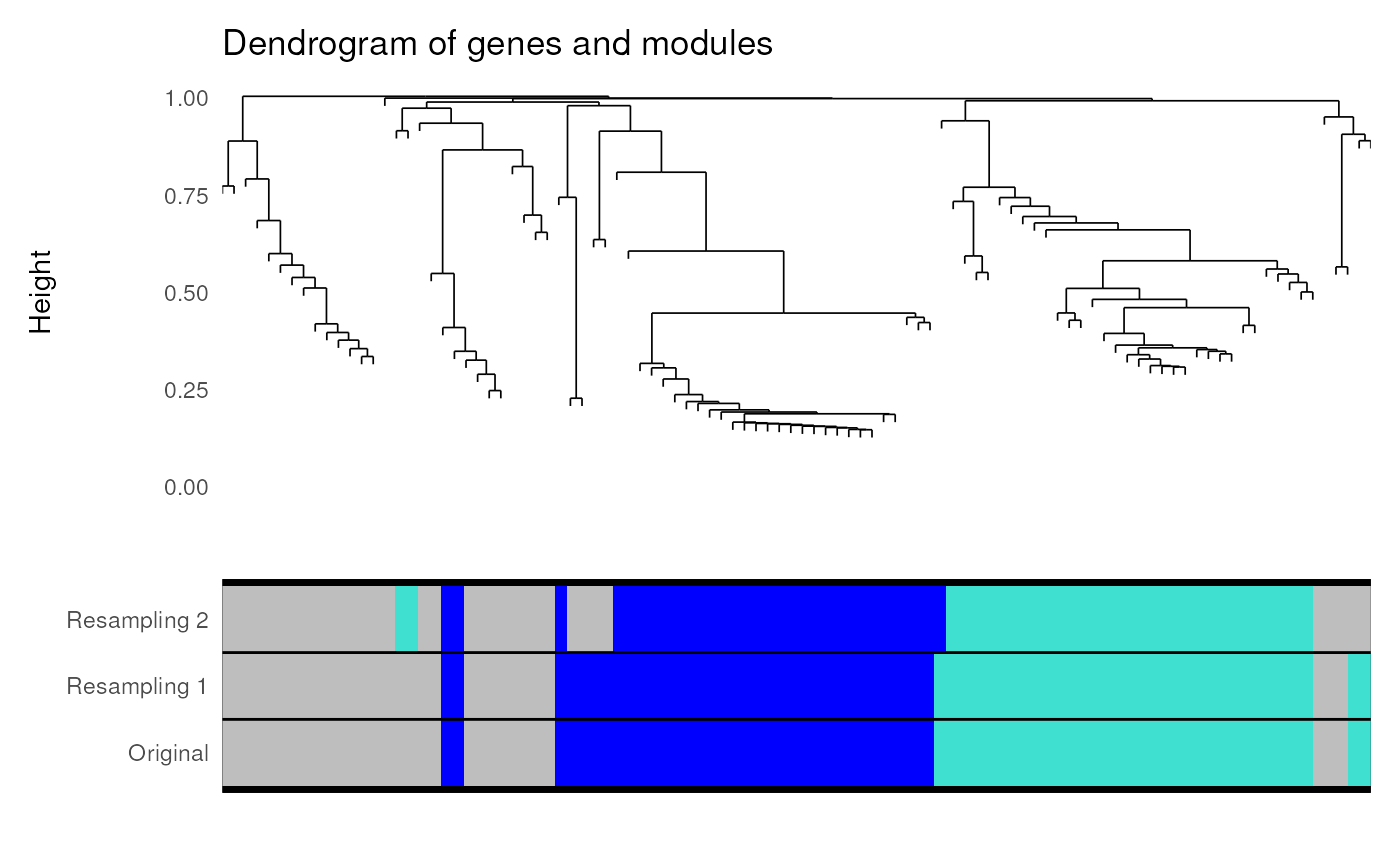
Perform module stability analysis
Examples
data(filt.se)
filt <- filt.se[1:100, ] # reducing even further for testing purposes
# The SFT fit was previously calculated and the optimal power was 16
gcn <- exp2gcn(filt, SFTpower = 16, cor_method = "pearson")
#> ..connectivity..
#> ..matrix multiplication (system BLAS)..
#> ..normalization..
#> ..done.
# For simplicity, only 2 runs
module_stability(exp = filt, net = gcn, nRuns = 2)
#> ...working on run 1 ..
#> ...working on run 2 ..
#> ...working on run 3 ..