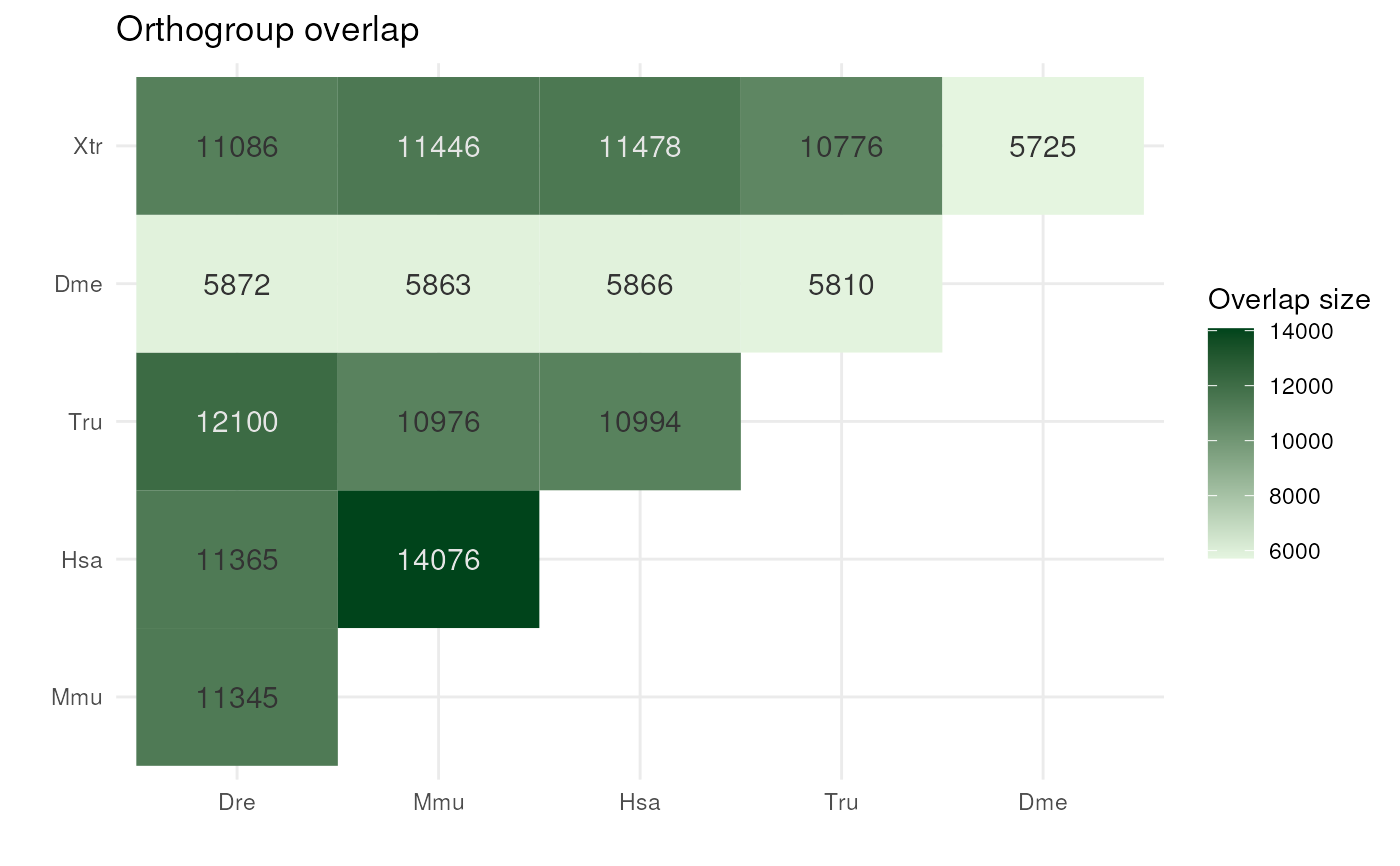
Plot pairwise orthogroup overlap between species
Arguments
- stats_list
A list of data frames with Orthofinder summary stats as returned by the function
read_orthofinder_stats.- clust
Logical indicating whether to clust data based on overlap. Default: TRUE
Examples
dir <- system.file("extdata", package = "cogeqc")
stats_list <- read_orthofinder_stats(dir)
plot_og_overlap(stats_list)