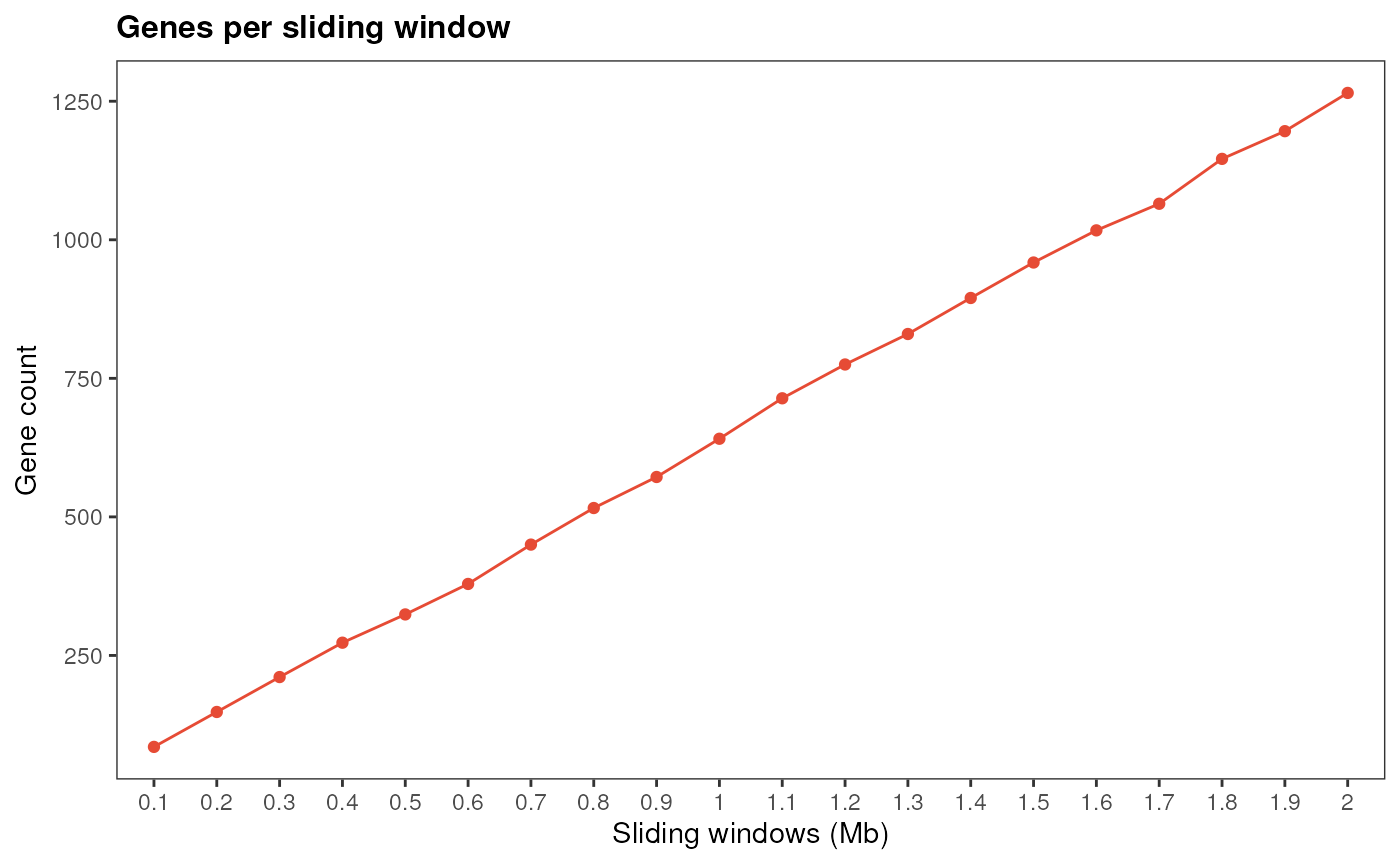
Simulate number of genes for each sliding window
Source:R/genomic_intervals_operations.R
simulate_windows.RdThis function counts genes that are contained in sliding windows related to each SNP.
Usage
simulate_windows(
gene_ranges,
marker_ranges,
windows = seq(0.1, 2, by = 0.1),
expand_intervals = TRUE
)Arguments
- gene_ranges
A GRanges object with genomic coordinates of all genes in the genome.
- marker_ranges
Genomic positions of SNPs. For a single trait, a GRanges object. For multiple traits, a GRangesList or CompressedGRangesList object, with each element of the list representing SNP positions for a particular trait.
- windows
Sliding windows (in Mb) upstream and downstream relative to each SNP. Default: seq(0.1, 2, by = 0.1).
- expand_intervals
Logical indicating whether or not to expand markers that are represented by intervals. This is particularly useful if users want to use a custom interval defined by linkage disequilibrium, for example. Default: TRUE.
Details
By default, the function creates 20 sliding windows by expanding upstream and downstream boundaries for each SNP from 0.1 Mb (100 kb) to 2 Mb.